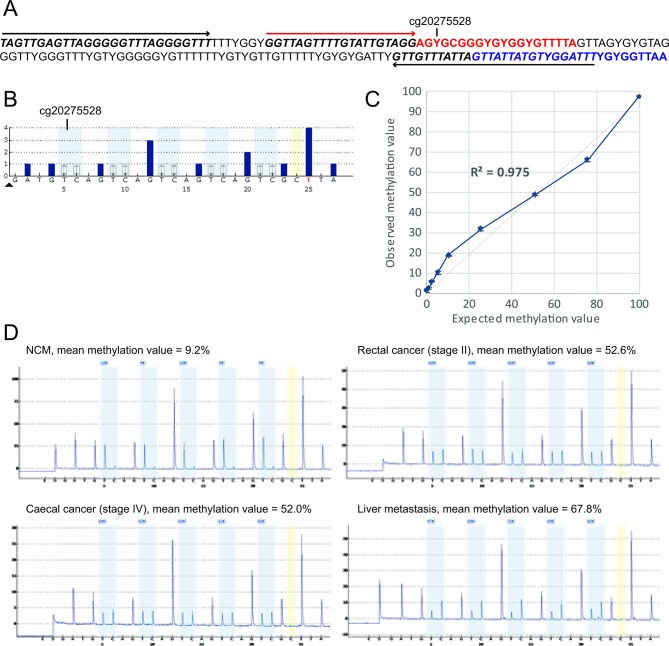
Figure 2.
Measurement of SEPTIN9 methylation levels by quantitative CpG pyrosequencing in archival Lynch syndrome (LS)-associated colorectal cancer (CRC), advanced adenoma, and paired normal colorectal mucosa (NCM) tissues. (A) Assay design. Sequence coordinates GRCh37/hg19 assembly Chr17: 75369414–75369587 are shown as bisulfite converted. YG indicates CpG sites where the cytosine may be methylated (CG) or unmethylated (TG). PCR primer-binding sites are italicised and indicated by black arrows. Pyrosequencing primer-binding site is italicised and indicated by a red arrow. The sequence analysed (red text) contains five CpG sites, of which the first corresponds to Infinium probe cg20275528, located 92 bp upstream of the fluorescent probe-binding sequence within the plasma-based Epi proColon 2.0 CE assay (blue text). (B) Simulated pyrosequencing assay. Grey shading shows the five CpG sites interrogated. Yellow shading shows the single cytosine control for bisulfite conversion efficiency. (C) Analytical sensitivity and linearity of the quantitative CpG pyrosequencing assay to measure methylation levels at SEPTIN9. The titration curve shows the observed (±1 SD) versus expected methylation values for the hypermethylated RKO CRC cell line DNA (expected value 100% methylated) diluted into the peripheral blood DNA from a healthy control (0% expected methylation value) in specific proportions. (D) Illustrative pyrograms obtained from formalin-fixed paraffin-embedded (FFPE) samples of primary CRC, a liver metastasis, and paired NCM from a Lynch syndrome patient with an MSH2 mutation.