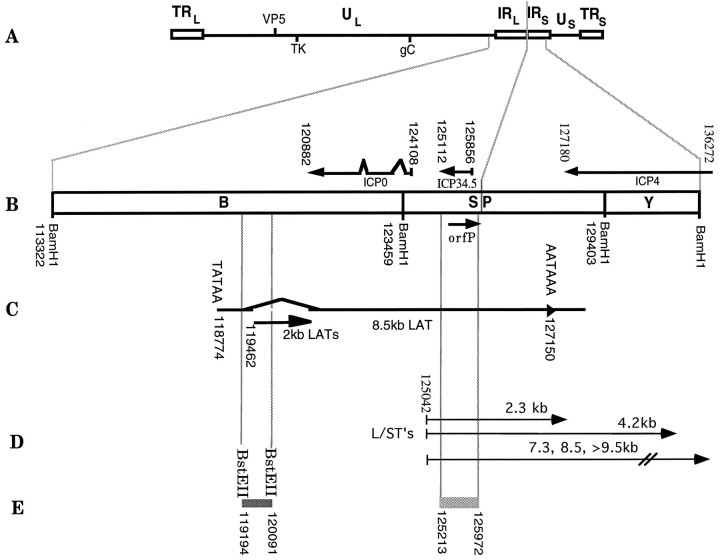
Fig. 1.
HSV-1 genome map. HSV-1 genome showing approximate location of the ICP34.5, 2 kb latency-associated transcripts (LATs), and neighboring genes. A, The 152 kb HSV-1 strain 17+ genome is shown that illustrates the unique long and short segments of the genome,UL and US(lines), bounded by internal (IR) and terminal repeat (TR) regions (open boxes). Hatch marks show location of the virion protein 5 (VP5), thymidine kinase (TK), and glycoprotein C (gC) genes. B, Expanded view of theUL/USregion of the genome showing the location of the ICP34.5, orfP, ICPO, and ICP4 genes. The transcripts in theBam SP region include the ICP34.5, ICP0, ICP4, mLAT, and L/STs. C, The location of the 2.0 kb LAT (2kb LATs) gene, which is expressed during acute and latent infection, and the primary 8.5 kb transcript (8.5kb LAT). D, The location of the proposed L/STs (Yeh and Schaffer, 1993).E, The location of the LAT-specificBstEII-BstEII probe used for in situ detection of HSV-specific latent gene expression and of one of the 759 bp deletions in strain 1716. Nucleotide positions are based on DNA sequence analysis of Perry and McGeoch (1988).