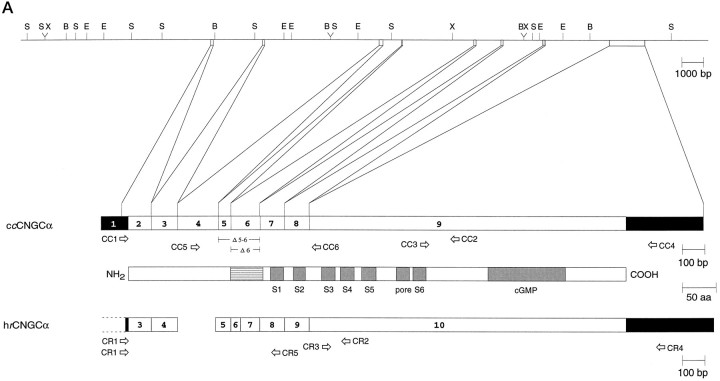
Fig. 1.
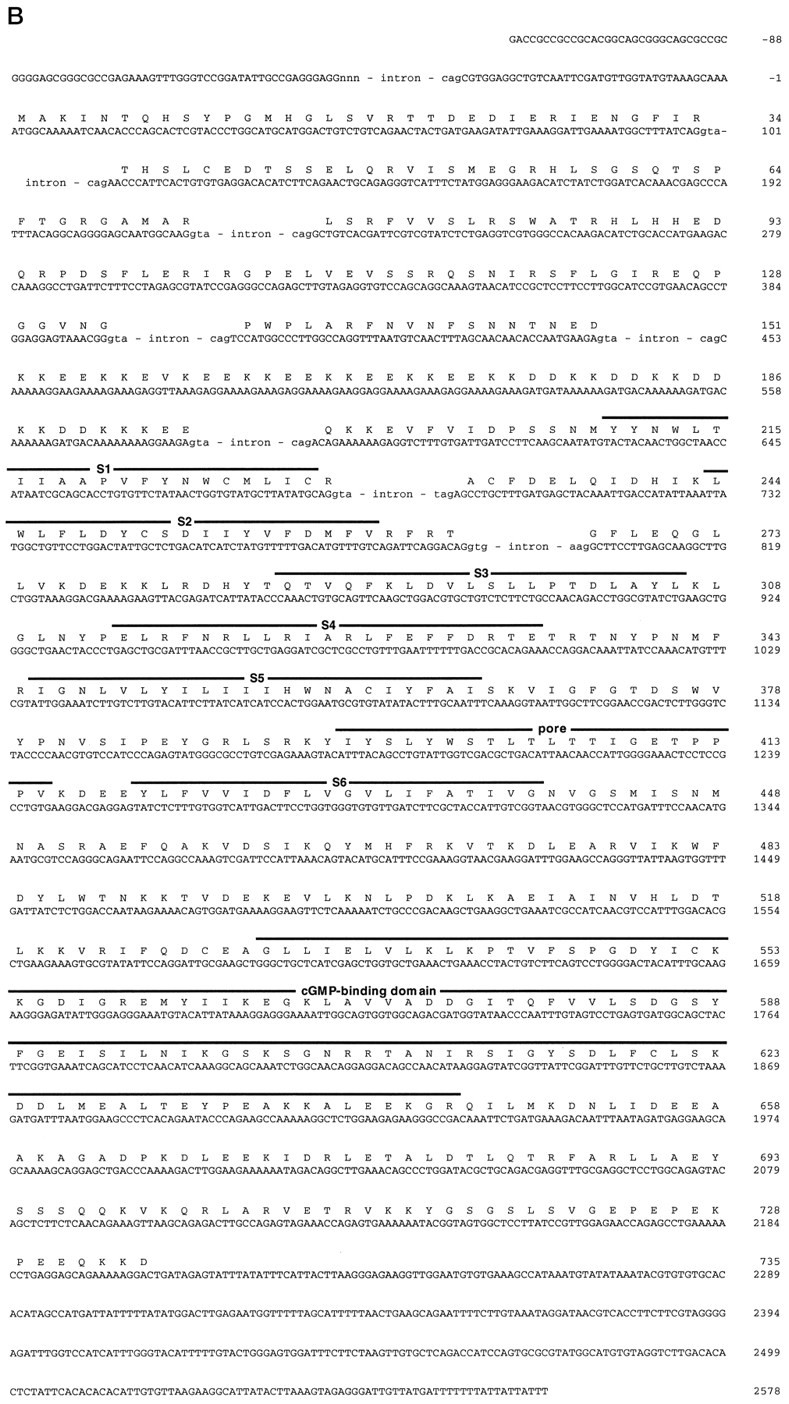
Gene structure of the chick cone CNG channel.A, Top, Restriction map of chromosomal DNA encompassing exons 1–9 of the chick cone CNG channel gene.B, BamHI; E,EcoRI; S, SacI;X, XbaI. A, Bottom, Comparison of the exon structure of the chick cone CNG channel (ccCNGCα) and the human rod CNG channel (hrCNGCα; Dhallan et al., 1992) with the transmembrane topology of CNG channels. Solid andopen boxes, respectively, refer to noncoding and coding regions of exons. Gray boxes refer to transmembrane segments S1–S6, the pore region, and the cGMP-binding site (cGMP); the striped box indicates the highly charged domain in the N-terminal region of the polypeptide. Thearrows refer to primer pairs CC1/CC2, CC3/CC4, and CC5/CC6 used for amplification of cone-specific cDNA fragments, or to the primer pairs CR1/CR2, CR3/CR4, and CR1/CR5 used for amplification of rod-specific cDNA fragments. B, Nucleotide sequence (with splice junction donor and acceptor sequences of exon/intron boundaries in the cDNA) and deduced amino acid sequence of ccCNGCα. Intron sequences at intron/exon boundaries are given in small letters. Numbering of nucleotide positions refers to the cDNA sequence and begins with +1 at the adenine nucleotide of the start codon. Figure continues.