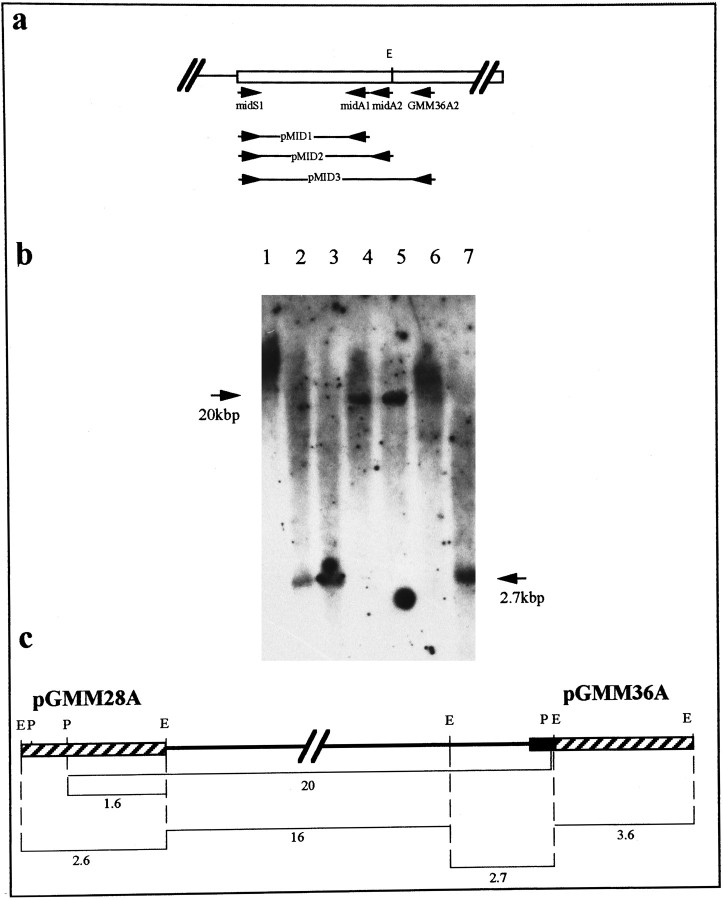
Fig. 3.
The genomic organization of the myomodulin gene.a, PCR amplification of the 5′ end of the second exon using primers specific for the cDNA sequence (arrows labeledGMM36A2, midS1, midA1, andmidA2) produced three products (pMID1, pMID2, and pMID3). Sequencing of these confirmed that the region of the cDNAs absent from genomic clones pGMM28A and pGMM36A is uninterrupted and lies at the 5′ end of the second exon. b, Southern blot analysis of restriction-digested Lymnaea genomic DNA using the PCR product pMID2 as a hybridization probe. Restriction digests with EcoRI (lanes 2, 3,7) produced a single band of 2.7 kb, whereas restriction digests with PstI (lanes 4,5) produced a 20 kb band (indicated with arrows). Because of the high-stringency washing of the filter, the secondPstI restriction fragment was not detected by the probe because of its short length of overlapping sequence (30 bp).1, Undigested DNA; 2, EcoRI;3, EcoRI/PstI; 4,PstI; 5, PstI/SalI;6, SalI; 7,EcoRI/SalI. This photograph has been electronically enhanced for improved clarity. c, Deduced map of the myomodulin gene with a single large intron of ∼19 kb (black line). All restriction fragment sizes are given in kb. The black region indicates the position of the PCR product pMID2 used as the hybridization in Figure 5B; the positions of genomic clones pGMM28A and pGMM36Aare indicated by diagonally striped boxes.