Abstract
Triple-negative breast cancers (TNBCs) lack progesterone and estrogen receptors and do not have amplified human epidermal growth factor receptor 2, the main therapeutic targets for managing breast cancer. TNBCs have an altered metabolism, including an increased Warburg effect and glutamine dependence, making the glutaminase inhibitor CB-839 therapeutically promising for this tumor type. Accordingly, CB-839 is currently in phase I/II clinical trials. However, not all TNBCs respond to CB-839 treatment, and the tumor resistance mechanism is not yet fully understood. Here we classified cell lines as CB-839–sensitive or –resistant according to their growth responses to CB-839. Compared with sensitive cells, resistant cells were less glutaminolytic and, upon CB-839 treatment, exhibited a smaller decrease in ATP content and less mitochondrial fragmentation, an indicator of poor mitochondrial health. Transcriptional analyses revealed that the expression levels of genes linked to lipid metabolism were altered between sensitive and resistant cells and between breast cancer tissues (available from The Cancer Genome Atlas project) with low versus high glutaminase (GLS) gene expression. Of note, CB-839–resistant TNBC cells had increased carnitine palmitoyltransferase 2 (CPT2) protein and CPT1 activity levels. In agreement, CB-839–resistant TNBC cells mobilized more fatty acids into mitochondria for oxidation, which responded to AMP-activated protein kinase and acetyl-CoA carboxylase signaling. Moreover, chemical inhibition of both glutaminase and CPT1 decreased cell proliferation and migration of CB-839–resistant cells compared with single inhibition of each enzyme. We propose that dual targeting of glutaminase and CPT1 activities may have therapeutic relevance for managing CB-839–resistant tumors.
Keywords: glutaminase, breast cancer, β-oxidation, energy metabolism, cancer therapy, CB-839, CPT1, CPT2, etomoxir
Introduction
TNBCs,3 which are characterized as estrogen receptor/progesterone receptor/HER2 negative tumors (1), do not respond to hormonal, monoclonal, and tyrosine kinase inhibitor therapies targeting the progesterone and estrogen receptors or Her2 (or its downstream signaling pathway). TNBCs have a worse prognosis, higher recurrence rate, and greater aggressiveness compared with other breast cancer subtypes (2, 3). Importantly, these tumors have a heterogeneous molecular profile, which hampers the discovery of biomarkers and more efficient therapies (4).
Metabolic reprogramming followed by increased glucose (and often glutamine) consumption is a hallmark of cancer (5) caused by the tumor cells' need to maintain high energy rates and biomass production. Metabolic reprograming also affects the migration and invasion processes (6). Recent studies have shown a relationship between TNBCs and altered metabolism (7–11), suggesting that metabolic reprogramming may be key to disease progression and a promising feature in the development of new therapies (12, 13). Specifically, glutamine is an important tricarboxylic acid cycle anaplerotic source for many types of tumors, including TNBCs (14–16). Glutamine catabolism affects tumor cell proliferation (17, 18), redox balance (14, 19), biosynthesis of other nonessential amino acids (20), and, importantly, maintenance of cancer stem cells (21); thus, it is highly linked to tumor recurrence (22).
The glutaminase (GLS) enzyme, which is involved in hydrolytic deamination of glutamine to glutamate and ammonium, is a limiting step in the glutamine catabolism process (23). The GLS gene generates two isoforms by alternative splicing, glutaminase C (GAC) and kidney-type glutaminase (KGA) (24). GLS inhibition has been explored as a therapeutic approach for different types of tumors (25–27), including TNBC (28). In fact, CB-839, a GLS inhibitor, is in phase I–II clinical trials for this type of breast cancer (29).
Structural lipids are synthesized de novo in cells when there is an energy surplus (16). Conversely, when the energy stock is low, fatty acids stored in triglycerides are released and catabolized by the β-oxidation process (30). The balance between lipid synthesis and catabolism is regulated by the energy sensor AMP-activated protein kinase (AMPK), which responds directly to intracellular AMP/ATP levels. When energy is low (high AMP/ATP levels), AMPK is activated and down-regulates fatty acid biosynthesis, with concurrent activation of mitochondrial β-oxidation (31).
β-Oxidation has been described as an essential energy source for TNBCs (32). It is also directly linked to cell aggressiveness (as measured by its effect on the migration and invasion processes) (33–35). Park et al. demonstrated that progression and metastasis in TNBCs are dependent on β-oxidation via c-Src activation and concluded that β-oxidation inhibition may be promising for TNBC patients (33).
Although it has generally been shown that TNBC depends on glutamine to survive, which is correlated with high GLS levels, it is clear that distinct cell lines (and tumors) respond differently to deprivation of this nutrient (14) and to GLS inhibition (28), suggesting a mechanism of resistance. We hypothesized that CB-839-resistant TNBC cells rely on nutrients other than glutamine to survive glutaminase inhibition. To evaluate this hypothesis, we characterized sensitive and resistant TNBC cell lines based on their response to CB-839 for cell proliferation. We then showed that resistant cell lines present lower GLS levels and increased β-oxidation (with a further increase upon CB-839 inhibition or GLS attenuation), a process that is linked to AMPK and ACC signaling and CPT1 activity. Breast tumors from a TCGA cohort with decreased GLS expression levels have increased CPT1A/B, CPT2, and carnitine O-acetyltransferase (CRAT) levels. Importantly, we showed that inhibiting both glutaminase and CPT1 decreased cell proliferation and migration. We propose that lower GLS levels combined with higher CPT1, CPT2, and CRAT levels may be a predictor of CB-839 resistance and that double GLS–CPT1 inhibition may be a promising treatment for TNBC.
Results
TNBC cell lines respond heterogeneously to glutamine withdrawal and glutaminase inhibition
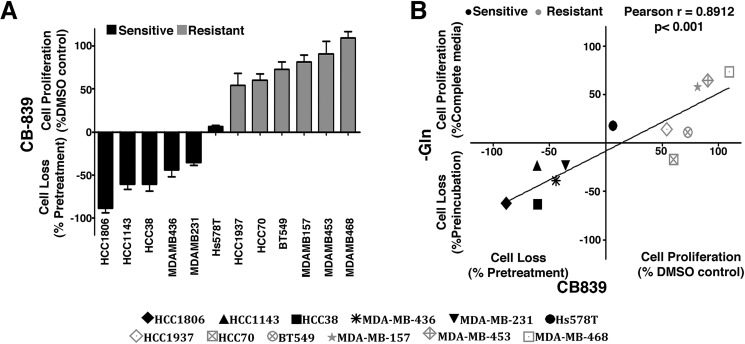
We evaluated 12 TNBC cell lines according to their sensitivity to glutaminase inhibition by CB-839 and glutamine dependence for cell proliferation. CB-839 treatment induced cell loss or decreased cell proliferation by more than 50% in six cell lines (HCC1806, HCC1143, HCC38, MDA-MB-436, MDA-MB-231, and Hs578T), which were then called sensitive cell lines; the other six cell lines (HCC1937, HCC70, BT549, MDA-MB-157, MDA-MB-453, and MDA-MB468) were either not affected or had their cell proliferation affected by less than 50% and were called resistant (Fig. 1A). CB-839 sensitivity was positively correlated with glutamine dependence for growth (Pearson correlation of 0.89) (Fig. 1B and Fig. S1A). To further characterize GLS inhibition dependence for growth, we determined the dose-dependent growth curve of bis-2-(5-phenylacetamide-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES), another GLS inhibitor (25), for three sensitive cells lines (MDA-MB-231, MDA-MB-436, and Hs578t) and three resistant cell lines (BT549, MDA-MB-468, and MDA-MB-453). Although the IC50 values of the sensitive cell lines ranged from 4 to 16 μm, the resistant cell lines displayed IC50 values between 36 and 74 μm (Fig. S1B).
Figure 1.
TNBC cell lines respond differently to glutaminase inhibition. A, growth response of 12 TNBC cell lines treated with 1 μm CB-839 for 4 days. The cell lines were classified as resistant when treatment affected growth in less than 50% (compared with the DMSO control) and sensitive when treatment affected growth in more than 50% (compared with the DMSO control) or led to cell death (when the number of cells at the end point was smaller than the number of seeded cells). B, correlation between the growth response after 4 days of CB-839 treatment or incubation of TNBC cells in glutamine-free medium. Data represent the mean ± S.D. of n = 4.
Resistant cell lines depend less on glutamine for mitochondrial function
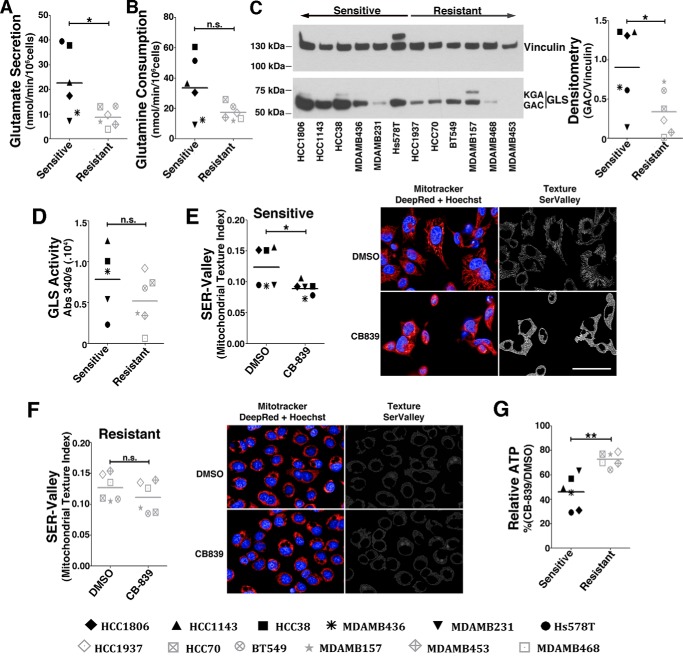
Glutamine addiction has been associated with high glutaminolytic rates (15) and elevated levels of the GLS protein, particularly the GAC isoform (14, 28). We evaluated the glutamine consumption, glutamate secretion, GAC protein levels, and glutaminase activity of the sensitive and resistant cell lines. As expected, the sensitive cell lines displayed increased glutamate secretion (Fig. 2A), a tendency for increased glutamine consumption (Fig. 2B), increased GAC levels (Fig. 2C), and a tendency of increased glutaminase activity (Fig. 2D) compared with resistant cell lines.
Figure 2.
Resistant cell lines are less glutaminolytic and use an alternative energy source. A and B, glutamate secretion (A) and glutamine consumption (B) of sensitive and resistant cell lines. C, Western blot displaying GLS (GAC and kidney-type glutaminase (KGA) bands are indicated) protein levels. Shown is densitometry of GAC to vinculin band intensity ratio. D, GLS activity of the whole-cell lysate of sensitive and resistant cell lines (the HCC1806 cell line was not evaluated). E and F, SER Valley texture classification (1 pixel) from SER Features of MitoTracker Deep Red–labeled mitochondria of sensitive (E) and resistant cell lines (F) treated with CB-839 (or DMSO control). Shown are representative images of the sensitive HCC1806 (E) or resistant MDA-MB-468 cells (F). Scale bar = 50 μm. Hoechst staining was performed for nucleus detection (blue). G, relative ATP levels in CB-839–treated cells represented as a percentage of vehicle treatment (DMSO). In the boxplots in A, B, and D–G, each point represents the mean of n = 4 of each cell line. Student's t test was applied. *, p < 0.05; **, p < 0.01; n.s., nonsignificant.
We then evaluated the effect of CB-839 treatment on the mitochondrial morphology of the sensitive and resistant cell lines. Tubular mitochondria are associated with network formation by the fusion process, which is indicative of healthy and functional organelles. However, a more granular structure can be associated with energy stress and the apoptotic process (36). Mitochondrial morphology was determined by the SER Valley texture parameter (37) using MitoTracker Deep Red staining. The higher the texture index, the more tubular (and more active) the mitochondria, whereas the opposite indicates less active mitochondria. Overall, upon CB-839 treatment, the sensitive cell lines had a heterogeneous but, on average, significant decrease in the texture index (Fig. 2E), whereas the resistant cell lines failed to respond to the treatment in a more homogenous manner (Fig. 2F). In accordance, CB-839 treatment (compared with DMSO) decreased the ATP levels of the sensitive cell lines by 54% but affected the resistant cell lines by only 27%; the difference between groups was significant (Fig. 2G). We conclude that resistant cell lines are less glutaminolytic (and less dependent on glutamine) and, upon GLS inhibition, have increased participation of metabolites other than glutamine in the tricarboxylic acid cycle for ATP production.
Resistant cell lines and tumors with low GLS levels present differential expression levels of lipid metabolism genes
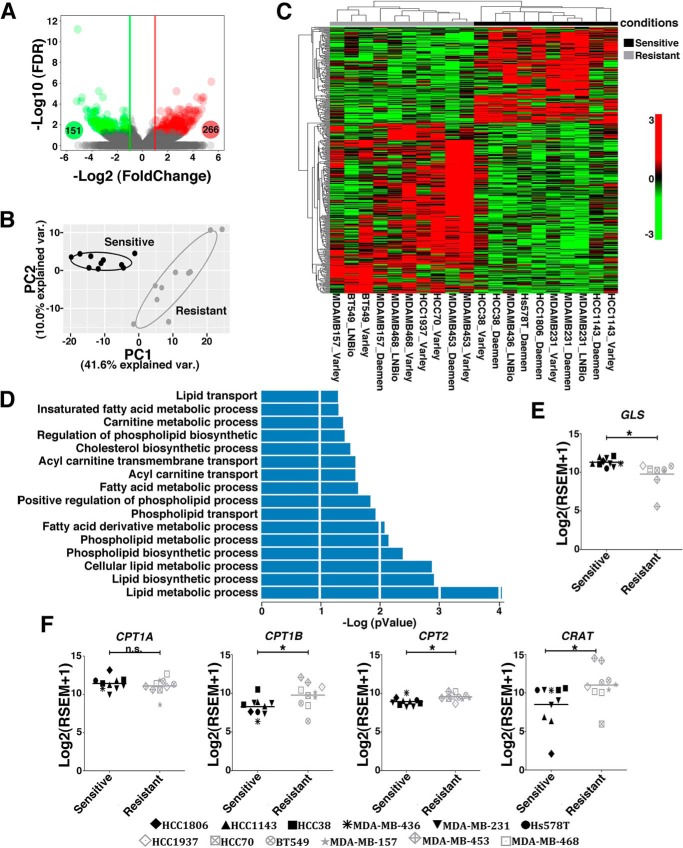
We previously performed an RNA-Seq analysis of TNBC cell lines (38). By using this dataset, we separated the cell lines into sensitive and resistant and determined the differentially expressed (DE) genes. Four hundred and seventeen genes were DE beyond the established cutoff (adjusted p < 0.05), with 266 being up-regulated (log2 -fold change (FC) ≥ +1) and 151 down-regulated (log2 FC ≤ −1) in the resistant cell lines (Fig. 3A). The DE genes clearly distinguish two groups, as determined by an unsupervised principal component analysis (Fig. 3B) and hierarchical clustering (Fig. 3C).
Figure 3.
Resistant and sensitive TNBC cell lines present gene expression alterations in lipid metabolism pathways. A, volcano plot showing DE (green circles, down-regulated, Log2FC ≤ −1; red circles, up-regulated, log2FC ≥ +1; false discovery rate ≤ 0.05) genes between resistant and sensitive cell lines. B and C, unsupervised principal component analysis (B) and heatmap clustering (C) performed with DE genes revealed a clear separation between resistant and sensitive cell lines. D, biological processes related to lipid metabolism obtained from the pathway enrichment analysis (p ≤ 0.05). E and F, boxplots presenting differences in the expression levels of GLS (E) and CPT1A, CPT1B, CRAT, and CPT2 (F) between sensitive and resistant cell lines. Twenty samples were used in this analysis, with some of the cell lines presented as replicates (obtained from our laboratory or from data banks (38). We evaluated the resistant cell lines MDA-MB-453 (2 samples), HCC1937 (1 sample), MDA-MB-468 (2 samples), HCC70 (1 sample), MDA-MB-157 (2 samples), and BT549 (2 samples) and the sensitive cell lines HCC1143 (2 samples), HCC38 (2 samples), MDA-MB-436 (1 sample), MDA-MB-231 (3 samples), Hs578 (1 sample), and HCC1806 (1 sample). Student's t test was applied. *, p < 0.05; n.s., nonsignificant.
We then performed an enrichment pathway analysis using the Gene Ontology (GO) database and found that, below the applied cutoff of p < 0.05, 16 biological processes related to lipid metabolism were enriched, among others (Fig. 3D, Table S1, and data not shown). These pathways contained 56 DE genes, with 44 genes up-regulated and 12 down-regulated in the resistant group (Table S2). Interestingly, 7 DE genes related to β-oxidation/lipid catabolism processes were up-regulated in resistant cell lines (Table S2). Altogether, these data suggest that the ability to metabolize lipids is associated with increased resistance to glutaminase inhibition.
CPT1A and CPT1B, CRAT, and CPT2 gene products are directly involved in the transport of fatty acids into mitochondria and serve as limiting steps for the β-oxidation process. In agreement with our hypothesis, we found that resistant cell lines, in addition to presenting lower GLS expression levels (Fig. 3E), present increased CPT1B (but not CPT1A), CRAT, and CPT2 expression levels compared with sensitive cell lines (Fig. 3F).
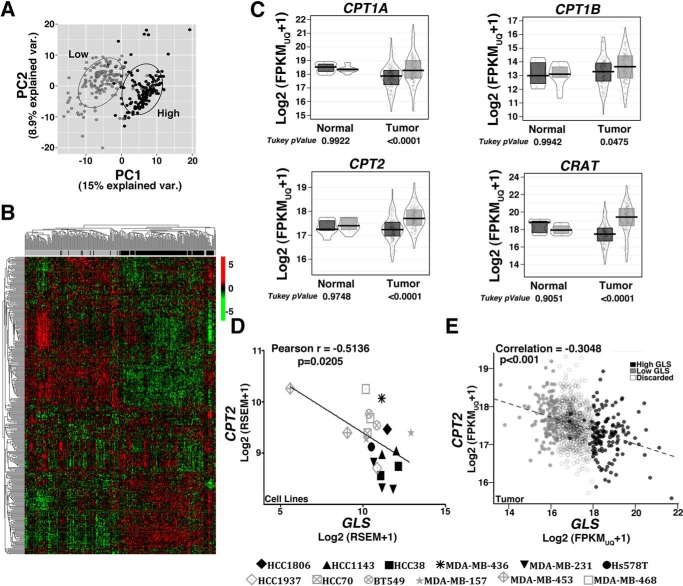
Subsequently, we analyzed RNA expression data from 1097 breast tumors (available from TCGA). Tumor tissues were classified based on GLS expression, and the 12.5% of cases with the highest (137 tumors, with GLS expression ranging from log2 FPKM-UQ+1 of 17.97 to 21.71) and the 12.5% of cases with the lowest (137 tumors, log2 FPKM-UQ+1 between 13.60 and 16.69) GLS mRNA levels were classified as the “high” or “low” GLS groups, respectively. Then, using 337 genes of the “fatty acid metabolic process” (GO 0006631), we performed an unsupervised principal component analysis and hierarchical clustering analysis to determine whether the high and low GLS groups differed from one another. Indeed, most of the tumors within the high or low GLS classifications were clustered together, whereas the groups were generally separated from one another (Fig. 4, A and B, respectively). We found that 270 genes of the GO “fatty acid metabolic process” were DE (adjusted p < 0.05), of which 162 were up-regulated and 108 were down-regulated in low GLS; among those, 75 genes were related to “fatty acid oxidation process” (intersection between GO 0006631 and GO 0019395), with 52 genes (70% of them) up-regulated in low GLS (Table S3). We also observed that, although CPT1A, CPT1B, CRAT, and CPT2 expression levels did not change in normal adjacent breast tissues, they were significantly more expressed in tumor tissues with low GLS levels (Fig. 4C). Moreover, there was a negative Pearson correlation between CPT2 and GLS expression levels in the evaluated cell lines (−0.5136; Fig. 4D) and breast tumor tissues (−0.3048; Fig. 4E). These results show that resistant cell lines and tumor tissues with low GLS levels present increased levels of genes related to fatty acid catabolism.
Figure 4.
Low and high GLS breast tumors present altered expression of lipid metabolism-related genes. A and B, unsupervised principal component analysis (A) and heatmap clustering (B) performed with the 337 genes related to “fatty acid metabolism process” (GO 0006631) show a separation between breast tumors classified as low or high GLS levels. C, boxplot presenting differences in the expression levels of CPT1A/B, CRAT, and CPT2 between low and high GLS levels in normal tissue and in breast tumors. D and E, correlation between CPT2 and GLS expression levels in cell lines (D) and breast tumors (E). In C, an ANOVA and Tukey's multiple comparisons were applied. In D, 20 samples were used for analysis, with some of the cell lines presented as replicates (obtained from our laboratory or data banks, (38). We evaluated the resistant cell lines MDA-MB-453 (2 samples), HCC1937 (1 sample), MDA-MB-468 (2 samples), HCC70 (1 sample), MDA-MB-157 (2 samples), and BT549 (2 samples) and the sensitive cell lines HCC1143 (2 samples), HCC38 (2 samples), MDA-MB-436 (1 sample), MDA-MB-231 (3 samples), Hs578 (1 sample), and HCC1806 (1 sample). In E, 1097 samples were used. Light gray, low-GLS samples; black, high-GLS samples.
Resistant cell lines present increased mitochondrial β-oxidation
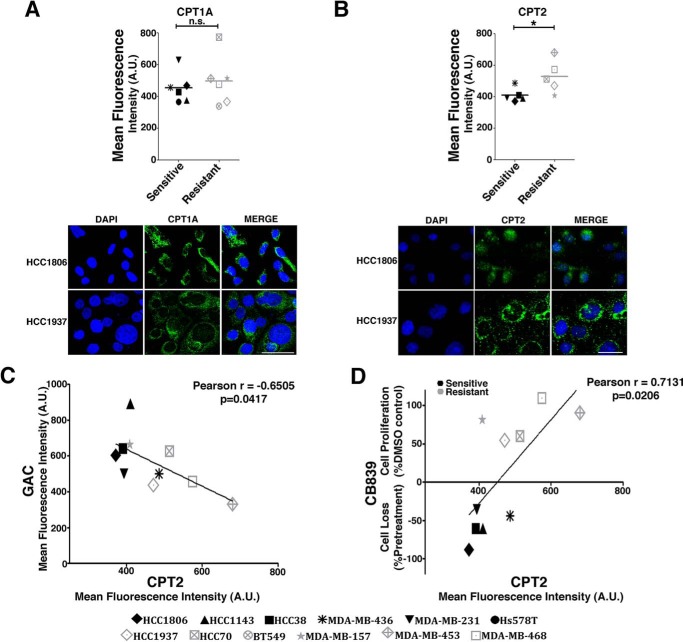
CPT1B and CPT2 expression levels were enhanced in the resistant cell lines but not CPT1A (although it was enhanced in breast tumor tissues with low GLS levels). CPT1A is a more ubiquitous isoform of CPT1, so we evaluated both CPT1A and CPT2 protein levels in resistant and sensitive cells. CPT2 protein levels (but not CPT1A levels) were, on average, increased in resistant compared with sensitive cell lines (Fig. 5, A and B). Moreover, there was a negative correlation between GAC and CPT2 protein levels (Pearson correlation of −0.6505; Fig. 5C). Concordant with CPT2 protein levels being related to CB-839 resistance, we observed a positive Pearson correlation between CPT2 protein levels and a decreased effect of CB-839 on cell proliferation and survival (Pearson correlation of 0.7131; Fig. 5D).
Figure 5.
CPT2 levels are negatively correlated with GAC and CB-839 resistance. A and B, CPT1A (A) and CPT2 (B) protein levels as quantified by immunofluorescence of sensitive and resistant cell lines. Representative images of the sensitive HCC1806 cell line and the resistant HCC1937 cell line are shown below. Scale bars = 50 μm. DAPI staining was used for nucleus detection. C and D, Pearson correlation coefficient between the CPT2 and GAC immunofluorescent signal (C) and CB-839 effect on growth and the CPT2 immunofluorescent signal (D). In B–D, 10 cell lines were evaluated (Hs578T and BT549 cell lines were not evaluated). In A and B, Student's t test was applied. *, p < 0.05; n.s., nonsignificant; A.U., arbitrary units.
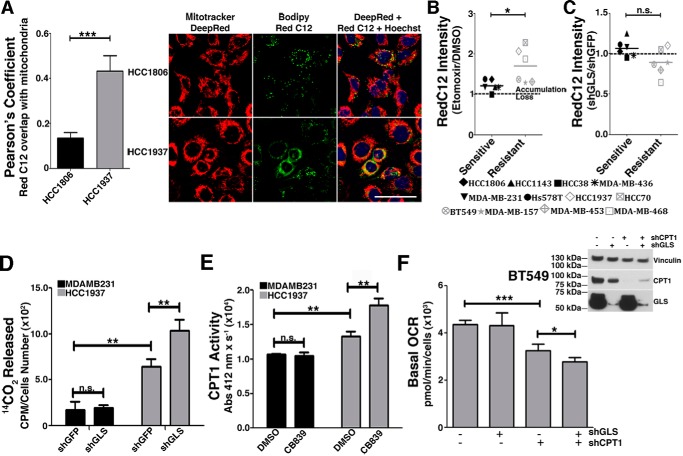
To evaluate FA entrance into mitochondria, a crucial step for the β-oxidation process, we measured the colocalization (displayed as the Pearson correlation) of fluorescence signals of the BODIPY 558/568 C12 (RedC12), and MitoTracker Deep Red probes (39). We verified that the resistant cell line HCC1937 presented higher signal colocalization of both probes than the sensitive cell line HCC1806 (Fig. 6A). We then evaluated the decrease of the RedC12 fluorescence signal in the resistant and sensitive cell lines, which is related to an increase in β-oxidation (39). We observed that resistant cell lines treated with etomoxir (a CPT1 inhibitor (40)) presented greater probe accumulation than sensitive cell lines (Fig. 6B). This result is in agreement with a scenario where resistant cells mobilize and likely consume more fatty acids in mitochondria through β-oxidation.
Figure 6.
Resistant TNBC cell lines have increased mitochondrial β-oxidation, which is further increased by GLS attenuation. A, Pearson correlation coefficient between BODIPY 558/568 C12 (RedC12) and MitoTracker Deep Red in the sensitive HCC1806 cell line and the resistant HCC1937 cell line. Representative images of the cell lines are shown on the right. Hoechst staining was performed for nucleus detection. Higher correlation implies increased co-localization between the FA probe and mitochondria. Scale bar = 50 μm. B, BODIPY accumulation caused by etomoxir treatment in sensitive and resistant cells lines. C, four (BT549, MDA-MB-157, MDA-MB-453, and MDA-MB-468) of the six resistant cell lines and two (HCC38 and MDA-MB-436) of the six sensitive cell lines responded to GLS knockdown by decreasing the RedC12 fluorescent signal compared with the shGFP control, implying increased β-oxidation in these cells when GLS is attenuated. D, β-oxidation was directly measured by quantifying 14CO2 released from uniformly labeled [14C]palmitic acid. The resistant HCC1937 cell line released more 14CO2 than the sensitive cell line MDA-MB-231, which was further increased by GLS knockdown only in the resistant cell line. E, CPT1 activity was higher in the resistant HCC1937 cell line compared with the sensitive cell line MDA-MB-231, which was further increased by GLS knockdown only in the resistant cell line. F, the Western blot on the right shows GLS and CPT1A knockdown efficiency. Shown on the left is the basal OCR of the resistant BT549 cell line after individual or combined knockdown of GLS and CPT1A. In B and C, each point represents the mean of n = 4 of each cell line. In D--F, each bar represents the mean ± S.D. of n = 3. Student's t test was applied. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., nonsignificant.
Next we measured the effect of knocking down GLS (Fig. S2A) (compared with control shGFP) on β-oxidation in both sensitive and resistant cell lines. We verified that the shGLS/shGFP ratio of the RedC12 fluorescence intensity was, on average, 1.07 in the sensitive cell lines (a 7% increase in probe accumulation upon GLS knockdown), whereas, in the resistant cells, the ratio was, on average, 0.89, implying an 11% decrease in probe accumulation and a potential increase in β-oxidation upon GLS knockdown. Differences between groups were nonsignificant (Fig. 6C).
To directly measure β-oxidation, we quantified 14CO2 released from the oxidation of [14C]palmitic acid and CPT1 activity in resistant and sensitive cell lines. We verified that, in complete medium supplemented with [14C]palmitic acid, the resistant cell line HCC1937 released more 14CO2 than the sensitive cell line MDA-MB-231 compared with control shGFP cells (Fig. 6D). Similarly, CPT1 activity was higher in DMSO-treated HCC1937 cells than in DMSO-treated MDA-MB-231 cells (Fig. 6E). We then evaluated the effect of GLS attenuation on 14CO2 release and CPT1 activity in the resistant MDA-MB-231 and sensitive HCC1937 cell lines. Strikingly, GLS knockdown led to a significant increase in 14CO2 release of HCC1937 cells (compared with control shGFP) but not in MDA-MB-231 cells (Fig. 6D). In accordance, CB-839 treatment increased the CPT1 activity of HCC1937 cells (compared with DMSO) but not of MDA-MB-231 cells (Fig. 6E). In addition, we verified that CB-839 treatment increased the 14CO2 release of the resistant BT549 and HCC1937 cell lines (compared with DMSO; Fig. S2B) and the CPT1 activity of resistant HCC70 and BT549 cell lines (Fig. S2C). GLS knockdown in BT549 cells also promoted an increase in CPT1 activity (compared with shGFP; Fig. S2D). Next we evaluated the effect of simultaneous knockdown of GLS and CPT1 on the basal oxygen consumption rate (OCR) of resistant BT549 cells. Although GLS knockdown did not affect the basal OCR, suggesting that alternative sources of carbon (other than glutamine) were being used for mitochondrial oxidation, CPT1 knockdown significantly decreased the OCR compared with shGFP (Fig. 6F). Knocking down both GLS and CPT1 led to a further decrease in OCR (compared with shCPT1), revealing that GLS attenuation makes cells more dependent on CPT1 for respiration (Fig. 6F). Taken together, these data show that resistant cells have increased mitochondrial β-oxidation, which is further stimulated by GLS attenuation.
Resistant cell lines rely on CPT1 for growth and migration
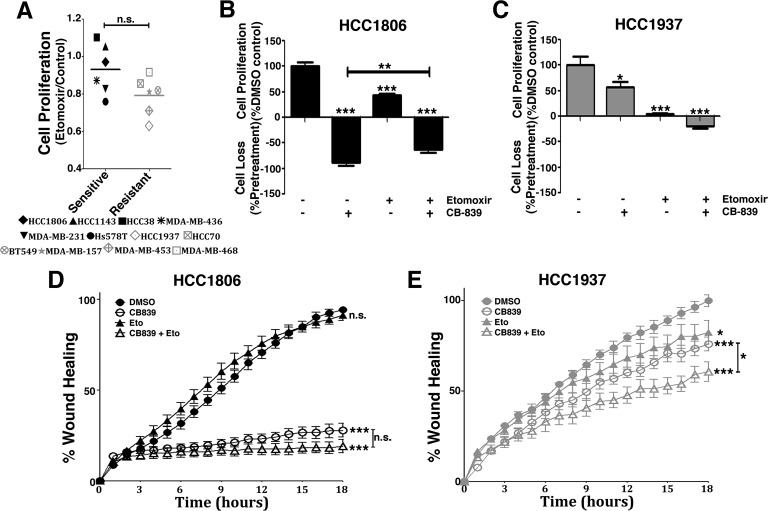
In agreement with resistant cell lines having increased CPT1 activity (and β-oxidation), we evaluated their growth dependence on CPT1. We first verified that these cells have a tendency of growing less in the presence of etomoxir than sensitive cells (nonsignificant; Fig. 7A). Although the sensitive cell line HCC1806 was affected by etomoxir (compared with the DMSO control, 57% growth reduction), combined etomoxir and CB-839 treatment decreased the efficacy of CB-839 (by decreasing cell death induced by CB-839; Fig. 7B). More strikingly, although treating the resistant cell line HCC1937 with either CB-839 or etomoxir only reduced proliferation (compared with the DMSO control, 43% and 95% decreases, respectively), a combination of both drugs led to cell death (20% cell loss; Fig. 7C).
Figure 7.
CB-839 treatment and CPT1 inhibition have a combined effect on the proliferation and migration of resistant cell lines. A, growth response of sensitive and resistant cell lines treated with etomoxir for 4 days. B and C, both sensitive HCC1806 (B) and resistant HCC1937 (C) cell lines responded to etomoxir treatment by decreasing cell growth, which was more pronounced in the resistant cell line. Combined CB-839 and etomoxir treatment led to cell death in the resistant cell line and, curiously, attenuated the effect of CB-839 in the sensitive cell line. D and E, combined CB-839 and etomoxir treatment did not further increase the effect of CB-839 treatment in the sensitive cell line HCC1806 (D) but had the additional effect on decreasing cell migration in the resistant cell line HCC1937 (E). In A, each point represents the mean of n = 4 of each cell line. In D and E, each bar represents the mean ± S.D. of n = 4. Student's t test was applied. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., nonsignificant. When not indicated, comparison was made against the DMSO control.
Glutamine metabolism by glutaminase and β-oxidation is already linked to the cell migration and invasion processes (41). We evaluated the effects of CB-839, etomoxir, and combined treatment on the migration of the HCC1806 and HCC1937 cell lines. In HCC1806 cells, CB-839 decreased cell migration by 70% (compared with the DMSO control) in 18 h, whereas etomoxir caused no pronounced effect alone (Fig. 7D). Etomoxir and CB-839 combination promoted a small and nonsignificant further decrease in cell migration in 18 h compared with CB-839 treatment alone (Fig. 7D). Conversely, HCC1937 cells responded similarly to either CB-839 or etomoxir (compared with DMSO, 24% and 18% decreases, respectively, in 18 h); combined treatment led to a significant further decrease compared with CB-839 alone, with a final 40% decrease in migration compared with the DMSO control (Fig. 7E). Similar results were observed in the BT549 and HCC70 resistant cell lines (Fig. S3, A and B, respectively). In summary, we conclude that CPT1 sustains the growth of resistant cell lines treated with CB-839. In addition, cell migration can be decreased in resistant cells upon double glutaminase and CPT1 inhibition.
GLS inhibition caused increased CPT1 activity and β-oxidation via AMPK pathway activation
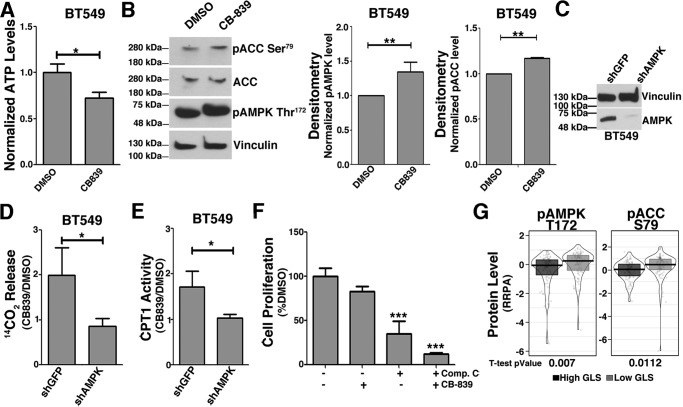
AMPK is phosphorylated on Thr-172 and activated when cells meet low energy levels. In turn, AMPK phosphorylates ACC on Ser-79, deactivating this enzyme and leading to a decrease in malonyl-CoA, a CPT1 inhibitor (42), which makes the AMPK pathway an obvious candidate for underlying the process of fatty acid catabolism linked to decreased glutaminase activity in TNBC. Indeed, glutaminase inhibition by CB-839 has been shown to induce AMPK activation in lung cancer cells (43).
Treating the resistant cell line BT549 with CB-839 decreased ATP (compared with DMSO; Fig. 8A) and increased AMPK Thr-172 and ACC Ser-79 phosphorylation levels (Fig. 8B). To further demonstrate AMPK involvement in CPT1 activation induced by GLS inhibition, we knocked down the AMPK α subunit (AMPKα) (Fig. 8C) and treated cells with CB-839. In shGFP cells, the CB-839/DMSO 14CO2 release ratio from [14C]palmitic acid was 2 (Fig. 8D); in agreement, the CB-839/DMSO CPT1 activity ratio was 1.7 (Fig. 8E). Conversely, in shAMPKα cells, GLS inhibition affected 14CO2 release and CPT1 activity levels only marginally compared with DMSO (CB-839/DMSO ratio of 0.9 and 1, respectively; Fig. 8, D and E, respectively). Finally, we verified that AMPK inhibition by compound C (44) decreased BT549 cell proliferation by 65% (compared with DMSO; Fig. 8F), an effect that was further enhanced to 89% with CB-839 co-treatment (compared with DMSO; Fig. 8F). Of note, we verified that breast tumor tissues with low GLS expression levels have increased total (data not shown) and phosphorylated ACC Ser-79 and phosphorylated AMPK Thr-172 levels compared with tissues with high expression of GLS (Fig. 8G). In summary, the AMPK pathway is important for the increase in CPT1 and β-oxidation triggered by GLS inhibition.
Figure 8.
GLS inhibition increases AMPK Thr-172 and ACC Ser-79 phosphorylation levels with a direct effect on β-oxidation. A, relative ATP levels in DMSO- and CB-839-treated BT549 resistant cells showing a 27% decrease with treatment. B, CB-839 treatment increases AMPK Thr-172 and ACC Ser-79 phosphorylation levels in BT549 cells related to total vinculin and ACC levels, respectively. C, Western blot showing AMPKα knockdown in BT549 cells. D and E, in BT549 cells, CB-839 treatment increases 14CO2 released from [14C]palmitic acid (D) and CPT1 activity (E) (compared with DMSO) in shGFP control cells but not in shAMPKα cells. F, proliferation of BT549 cells after CB-839 and compound C (an AMPK inhibitor) individual or combined treatment, showing that the resistant cell line relies on AMPK signaling for growth after glutaminase inhibition. G, breast tumor tissues with low GLS expression levels have increased AMPK Thr-172 and ACC Ser-79 phosphorylation levels compared with tissues with high GLS levels. The data in A and D–F represent the mean ± S.D. of n = 3. The data in B represent the mean ± S.D. of n = 4 (pAMPK) and n = 2 (pACC) independent western blots. The boxplot in G represents 111 tumors with high and 111 with low GLS gene expression, obtained from the reverse-phase protein array (RPPA); these samples overlap with the breast tumor samples obtained from the TCGA presented previously in the transcriptomics analysis. Student's t test was applied. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
In recent years, it has been shown that TNBCs depend on glutamine for growth and survival (14, 15, 45, 46). Glutamine and glutaminase are also involved in the gain of invasive traces in other tumor types (16, 47, 48). However, it is also clear that glutamine dependence varies within TN tumors, with some cell lines only marginally affected, whereas others stop growing or die via apoptosis after glutamine withdrawal (14) or glutaminase inhibition by CB-839 (28). Phase I and II clinical trials are being conducted with CB-839 for several solid (including TNBCs) and hematological tumors. Although glutaminase inhibition by CB-839 has advanced in the clinical trials, many studies are now being conducted in combination with other drugs. Indeed, enhanced performance was obtained when CB-839 was combined with β-lapachone, a compound that generates reactive oxygen species in cells, for treating pancreatic cancer (49); with a BCL-2 inhibitor to treat leukemia (50); and with paclitaxel, a cytoskeletal drug that targets tubulin, to treat TNBC (51). This can be explained by differences between molecular profiles and tumor microenvironments, which can generate context-dependent sensitivity to glutaminase inhibition (52). Metabolic plasticity is another factor that can drive CB-839 resistance.
In this work, we evaluated a set of TNBC cell lines for their relative sensitivity to CB-839 for growth. We defined resistant cell lines as those in which CB-839 only promoted growth inhibition of less than 50% (compared with vehicle); sensitive cell lines were defined as those that either grew less than 50% or died upon treatment. Although sensitive cells presented heterogenous behavior regarding glutamine metabolism, the evaluated resistant cell lines more homogenously presented decreased glutaminolysis and GLS levels. In addition, the resistant cells were less responsive than the sensitive cells to decreases in ATP when the cells were treated with CB-839. These results led us to speculate that nutrients other than glutamine were being metabolized for energy production in these cells upon glutaminase inhibition. By comparing the transcriptome of resistant and sensitive cells, we detected pathways linked to lipid metabolism that were altered between these cells. Specifically, we found that key genes for mitochondrial β-oxidation (CPT1B, CRAT, and CPT2) have increased expression in resistant cell lines. Strikingly, we also verified that tumor breast tissues separated into the 12.5% with the highest levels and the 12.5% with the lowest levels (called high GLS and low GLS, respectively) discriminated between groups as a role of the expression of genes related to the fatty acid metabolism GO process. Low versus high GLS–level tumor tissues also presented increased phosphorylation levels of the energy sensor AMPK (Thr-172) and its downstream target ACC (Ser-79). We concluded that GLS levels in breast tumor tissues were potentially related to changes in the lipid metabolism pathways. The mechanistic link between the expression levels of GLS and these genes deserves further investigation.
β-Oxidation has been connected to proliferation, migration, and invasion processes in TNBCs (32, 33, 35). Moreover, this process is related to metabolic adaptation under conditions of nutrient and oxygen deprivation in diverse tumor types (53, 54). More specifically, a study showed that withdrawal of glutamine from medium causes increases in proteins related to β-oxidation (55). Altogether, this information led us to speculate that β-oxidation is increased in resistant cell lines.
Indeed, we saw that resistant cells have increased CPT2 levels, mobilized more of a FA fluorescent probe (RedC12) to mitochondria, produced more 14CO2 from labeled palmitate, and presented increased CPT1 activity levels compared with sensitive cells. Resistant cells also degraded more RedC12 when GLS was knocked down (with sensitive cells, for some reason, accumulating the probe in this situation) and responded to CB-839 by increasing 14CO2 production and CPT1 activity.
Although we showed that resistant cell lines have an enhanced capacity to mobilize fatty acids for β-oxidation, we verified that the sensitive cell line MDA-MB-231 was also able, upon GLS knockdown, to increase the uptake of fatty acids from medium (measured by the C1-BODIPY C12 probe, Fig. S4A) to form more lipid droplets (data not shown) and to mobilize more neutral lipid droplets (quantified with a neutral lipid–specific dye) to lysosomes (Fig. S4B) than shGFP control cells. Upon transmission EM analysis, we confirmed the presence of cytoplasmic lipid droplets in these cells and their fusion to membrane-coated vesicles (Fig. S4C). However, unlike resistant cells, MDA-MB-231 did not respond to GLS knockdown by enhancing either 14CO2 or CPT1 activity levels (Fig. 6, D and E, respectively), showing no increase in the mitochondrial β-oxidation process. In fact, it was recently demonstrated that MDA-MB-231 cells respond to GLS inhibition (by another glutaminase inhibitor called C.968) by increasing autophagy, which increased cell survival upon glutaminase inhibition (56). Although the levels of metabolites related to lipid catabolism were altered upon treatment, the OCR induced by palmitate (the only direct evidence of β-oxidation in this work) indicated that cells could oxidize FAs; however, no data regarding the effect of GLS inhibition on this parameter was shown (56). In accordance, our data show that GLS attenuation by knockdown in this cell line is likely related to increased lipophagy. However, we failed to detect a direct increase in mitochondrial β-oxidation in this cell line induced by GLS attenuation, indicating that MDA-MB-231, although very responsive to CB-839 (and classified by us as sensitive), still presents a certain level of resistance in which lipid catabolism by mitochondrial β-oxidation may not be a relevant mechanism.
In addition, although GLS knockdown led to an increase in pAMPK Thr-172 and pACC Ser-79 in both MDA-MB-231 and the resistant cell line BT549 (data not shown for MDA-MB-231), only in the latter was this converted to increased CPT1 activity levels. This capacity of increasing CPT1 activity may be key to the metabolic adaptation process described here, and determining why resistant cells have an increased ability to do this (compared with sensitive cells) deserves further investigation.
Finally, we discovered that double glutaminase and CPT1 inhibition of resistant cells potentiates cell death and further decreased cell migration compared with individual treatments. Very importantly, in recent work, Yao et al. (57) showed that CPT1 has important metabolic roles related to cell proliferation that are independent of fatty acid oxidation. When working with etomoxir doses as low as 10 μm, they could measure a decrease in β-oxidation; however, this dose had no effect on proliferation (57). At higher doses, they showed that etomoxir has an off-target effect and can inhibit cell proliferation by mechanisms other than blocking β-oxidation (57). In our work, we used doses higher than 10 μm, and although we also measured a decrease in CPT1 activity and in β-oxidation, we cannot affirm that β-oxidation is directly responsible for the measured effects on proliferation and migration. In this regard, it is still an open question how exactly inhibiting CPT1 can correlate with GLS inhibition to further decrease proliferation (and migration) of resistant TNBC cells.
Finally, we propose that lower GLS levels associated with increased CPT1, CPT2, and CRAT mRNA levels can be potential markers to identify and select TNBCs that are poor CB-839 responders. This type of tumor may, from a clinical perspective, benefit from double glutaminase and β-oxidation inhibition. However, validation of these findings requires further in vivo proof.
Experimental procedures
Cell culture
HCC1806 (ATCC CRL-2335), HCC1143 (ATCC CRL-2321), HCC38 (ATCC CRL-2314), MDAMB436 (ATCC HTB-130), MDA-MB-231 (ATCC HTB-26), Hs578T (ATCC HTB-126), HCC1937 (ATCC CRL-2336), HCC70 (ATCC CRL-2315), BT549 (ATCC HTB-122), MDA-MB-157 (ATCC HTB-24), MDA-MB-453 (ATCC HTB131), and MDA-MB-468 (ATCC HTB-132) cells were maintained in RPMI 1640 medium supplemented with 10% FBS and incubated at 37 °C under 5% CO2 in a humidified atmosphere. All cell lines were obtained from the ATCC.
Lentiviral shRNA cloning and subcell line generation
pLKO.1 puro was a gift from Bob Weinberg (Addgene plasmid 8453) (58). Lentiviral shRNAs targeting the genes of interest were cloned in pLKO.1 within the AgeI/EcoRI sites at the 3′ end of the human U6 promoter. The targeted sequences were as follows: GFP, 5′-CAAGCTGACCCTGAAGTTCAT-3′; GLS, 5′-CAACTGGCCAAATTCAGTC-3′; CPT1, 5′-CGATGTTACGACAGGTGGTTT-3′; AMPKα, 5′-ATGAGTCTACAGCTATACCAA-3′. The cell lines were transduced with lentiviral particles from the pLKO.puro shGLS, tet-pLKO.G418 shCPT1, or tet-pLKO.puro shAMPKα vectors. The subcell lines were maintained with 1000 μg/ml G418 (Sigma-Aldrich) (shCPT1) or 1 μg/ml puromycin (Life Technologies) (shGLS and shAMPKα). To induce knockdown in transduced cells with the tet-pLKO vector, we utilized 50 ng/ml doxycycline for 7 days.
Proliferation assay
The cells were seeded at a density of 62.5 cells/mm2 in 96-well plates in complete medium. For the glutamine deprivation assay, after 24 h, the medium was replaced with either complete or glutamine-free RPMI medium, both supplemented with 10% dialyzed FBS (Thermo Fisher). For the inhibition assays, the cells were incubated with complete medium added of vehicle (0.1% (v/v) DMSO), 1 μm CB-839 (Sigma-Aldrich), 50 μm etomoxir (Cayman), or 3 μm compound C (Sigma-Aldrich). Double-inhibition assays were performed with 1 μm CB-839 and 200 μm etomoxir (individually or in combination). The medium was replaced every 48 h, and the cells were fixed with 3.7% formaldehyde and stained with 0.5 μg/ml DAPI after 96 h of treatment (T1). A mirror plate was set for every experiment, and the cells were fixed 24 h after seeding (T0). The number of stained nuclei was quantified using an Operetta fluorescence microscope (PerkinElmer Life Sciences) plate reader and Columbus software (PerkinElmer Life Sciences). Cell proliferation (when the number of cells in T1 > the number of cells in T0) and cell loss (when the number of cells in T1 < the number of cells in T0) were calculated using the following equations: cell proliferation = {100 × [(T1compound/T0compound)/(T1DMSO/T0DMSO)]}; cell loss = {100 × [1- (T1compound/T0Compound)]}. The bis-2-(5-phenylacetamide-1,3,4-thiadiazol-2-yl)ethyl sulfide IC50 value for cell proliferation was determined after 48 h of incubation. Sigmoidal curve and IC50 values were calculated with GraphPad Prism 8.0.0 software.
Glutamine consumption and glutamate secretion
The assay was performed using a method published previously (59) with some modifications. Briefly, the cells were seeded at a density of 937.5 cells/mm2 in 96-well plates in 50 μl of RPMI complete medium and sat for 12 h. Next, 10 μl of medium was combined with 190 μl of a solution containing 50 mm Tris acetate (pH 8.6), 0.2 mm EDTA (pH 8.0), 2 mm NAD+, 50 mm dipotassium phosphate, and 0.3 units of l-glutamate dehydrogenase (Sigma-Aldrich); the absorbance was measured at 340 nm using an EnSpire plate reader (PerkinElmer Life Sciences). Then, 60 nm of recombinant glutaminase C (purified as described in Ref. 60) was added to the same reaction to obtain the total amount of glutamine. The glutamate and glutamine concentrations were estimated based on the slope of a standard curve. Data were normalized by the number of cells, which was calculated as described above.
GLS activity assay
This assay was performed using a method published previously (61) with some modifications. Cells seeded at a density of 2500 cells/mm2 in 60-mm dishes were lysed in a solution containing 150 mm sodium chloride, 25 mm HEPES (pH 8.0), 1 mm EDTA, and 0.01% Triton X-100. The cells were then added to 10 mm sodium pyrophosphate, 20 mm sodium fluoride, 10 mm sodium orthovanadate, 1 mm PMSF, 10 mm β-glycerophosphate, 10 μm leupeptin, 1 μm pepstatin, 2 μg/ml aprotinin, and 4 mm benzamidine, followed by 20 strokes through a 26-gauge needle. After that, the samples were quantified by the Bradford method (62). Ten micrograms of cell lysate was combined with 50 mm Tris acetate (pH 8.6), 0.5 units of bovine l-glutamate dehydrogenase, 2 mm NAD+, 20 mm dipotassium phosphate, and 3.5 mm l-glutamine in a 96-well plate. The absorbance at 340 nm was measured over time on an EnSpire plate reader (PerkinElmer Life Sciences), and the slope of the curve was used to measure glutaminase activity.
Western blotting
Experiments were performed as described previously (38). The following antibodies were used: anti-GLS (Abcam, ab156876), anti-AMPK (Cell Signaling Technology, 2532), anti-pAMPK Thr-172 (Cell Signaling Technology, 2535), anti-ACC (Cell Signaling Technology, 3662), anti-pACC Ser-79 (Cell Signaling Technology, 3661), anti-vinculin (ab18058), anti-actin (ab3280), and anti-CPT1A (Abcam, ab128568). Two anti-rabbit secondary antibody HRP-linked were used: one from Cell Signaling Technology (7074) at a 1:1000 dilution, and another one from Sigma-Aldrich (A0545) at a 1:5000 dilution. An anti-mouse secondary antibody from Sigma-Aldrich (A4416) was used at a 1:5000 dilution.
ATP measurement
The cells were treated with 1 μm CB-839 (or vehicle DMSO) for 48 h and then seeded at a density of 62.5 cells/mm2 in a 384-well white plate. After 24 h, the assay was performed with the CellTiter-Glo Luminescent Cell Viability Kit (Promega) according to the manufacturer's instructions.
Mitochondrial texture index
The cells were treated with 1 μm CB-839 (or vehicle DMSO) for 48 h and then seeded at a density of 187.5 cells/mm2 in a 96-well plate. After 24 h, the cells were incubated with 100 nm MitoTracker Deep Red (Thermo Fisher) and 2.5 μm Hoechst (Thermo Fisher) in RPMI medium without phenol red and 1% FBS for 45 min. Then the cells were washed once and maintained in complete medium supplemented with 1% FBS. Images were taken immediately using an Operetta microscope (PerkinElmer Life Sciences). The analysis was performed with Columbus software (PerkinElmer Life Sciences). The SER valley texture classification (1 pixel) from Saddles, Edges, Ridges (SER) Features was used to evaluate mitochondrial morphology. Higher index values are related to a more complex and active mitochondrial network.
Immunofluorescence microscopy
The cells were seeded at a density of 187.5 cells/mm2 in a 96-well CellCarrier plate (PerkinElmer Life Sciences). After 24 h, the cells were fixed with 3.7% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 5 min. The cells were then incubated with blocking solution (5 mg/ml sodium heparin (5000 IU/ml), 5 mg/ml dextran sulfate, 0.1% Tween 20, and 0.05% sodium azide) for 30 min, followed by incubation with blocking/permeabilization solution (1% BSA, 0.1% Triton X-100, 50 mm glycine, and 10% goat serum) for 1 h using a humidity chamber. Then the cells were washed three times with working solution (diluted 5 times with blocking/permeabilization solution) and incubated overnight at 4 °C with the primary antibodies anti-CPT2 (1:100, Abcam, ab181114), anti-CPT1A (1:500, Abcam, ab128568), or anti-GAC (1:300, Rheabiotech) diluted in work solution. Twelve hours later, the cells were washed three times with working solution and incubated for 2 h at room temperature with the secondary antibodies Alexa 488 rabbit (1:200, Invitrogen, A11008), Alexa 488 mouse (1:400, Invitrogen, A11017), or Alexa 633 rabbit (1:300, Invitrogen, A21070), respectively, and diluted in work solution. The cells were then washed three more times with working solution and incubated with 1 μg/ml DAPI for 10 min. Images were obtained using an Operetta microscope (PerkinElmer Life Sciences). The analysis was performed with Columbus software (Perkin Elmer Life Sciences).
Bioinformatic analysis of transcriptomics and proteomics data
RNA-Seq analysis of the TNBC cell lines HCC1806, HCC1143, HCC38, MDA-MB-436, MDA-MB-231, Hs578T, HCC1937, HCC70, BT549, MDA-MB-157, MDA-MB-468, and MDA-MB-453 was performed as described previously (38) using three data sources (38, 63, 64). The cells were separated into CB-839–sensitive and –resistant cells and were analyzed in groups. Differential gene expression among these groups was performed using the DESeq2 package (R statistical software) (65). Gene enrichment using Gene Ontology (66) was performed using goseq (67). Patient RNA-Seq gene expression data were downloaded from the Genomic Data Commons portal (https://portal.gdc.cancer.gov/).4 Invasive breast carcinoma cases (1097 samples) were separated according to high and low GLS levels as described under “Results.” A differential gene expression analysis between the high and low GLS groups using raw transcript counts was performed with DESeq2. Reverse-phase protein array data were downloaded from The Cancer Proteome Atlas portal (https://tcpaportal.org/tcpa/)4 as replicate-based normalized values (68).
CPT1 activity assay
CPT1 activity was measured according to a protocol published previously (69) with modifications. Cells seeded at a density of 2500 cells/mm2 in 60-mm dishes were lysed in a solution containing 100 mm Tris-HCl (pH 8.0), 0.1% Triton X-100, 10 mm sodium pyrophosphate, 20 mm sodium fluoride, 10 mm sodium orthovanadate, 1 mm PMSF, 10 mm β-glycerophosphate, 10 μm leupeptin, 1 μm pepstatin, 2 μg/ml, aprotinin and 4 mm benzamidine, followed by three cycles of freezing in dry ice and thawing in ice and then 20 strokes through a 26-gauge needle. The samples were quantified by the Bradford method (62). The lysates (25 μg) were combined with 2.25 mm 5,5′-dithiobis(2-nitrobenzoic acid) (Sigma-Aldrich) in a 384-well plate and incubated at 37 °C for 30 min. Then 100 μm palmitoyl-CoA (Sigma-Aldrich) and 5 μm l-carnitine (Sigma-Aldrich) were added to each well to reach a final volume of 50 μl. Absorbance at 412 nm was measured in an EnSpire plate reader (PerkinElmer Life Sciences), and the slope of the curve was used as the protein activity measurement.
Measurement of 14CO2 release
β-Oxidation was measured according to a protocol described previously (70) with modifications. The cells were seeded at a density of 2000 cells/mm2 in 12.5-cm2 flasks without filter caps. After 24 h, the cells were washed and maintained in 2.5 ml of RPMI medium (without sodium bicarbonate, glutamine, and glucose) supplemented with 2.5 mm glucose, 1 mm glutamine, 1 mm carnitine, 25 mm HEPES, 1% FBS, and 100 μm BSA–palmitate containing 0.1 μCi/ml of [14C]palmitic acid labeled uniformly (Amersham Biosciences, GE Healthcare; specific activity of 57 mCi/mmol). A Whatman filter paper soaked with 30 μl of 2 m potassium hydroxide was placed on the flask cap. After incubation at 37 °C for 3 h, the filters were placed in 1 ml of scintillation liquid, and the signal was measured using Beckman Coulter LS6500 multipurpose scintillation counter equipment (Beckman). To prepare 2.5 mm BSA–palmitate complexes, 7.5% BSA was dissolved in water at 37 °C to complete dilution. 76 mmol of sodium palmitate (Sigma-Aldrich) was mixed in water and held at 70 °C to complete the dilution. After that, the solutions were combined (162.5 μl of sodium palmitate and 4675 μl of BSA) and stirred at 37 °C until completely solubilized.
Measurement of basal OCR
The basal oxygen consumption rate was measured using Seahorse XFe24 Analyzer equipment according to the manufacturer's recommendations. Briefly, we seeded 937.5 cells/mm2 on the XF24 microplate with RPMI medium for 16 h. Then we replaced the RPMI medium with medium without FBS and sodium bicarbonate and incubated the plate for 1 h while calibrating the equipment. Then the plate was placed into the analyzer and the reading was performed. The number of cells was used to normalize the data.
Fluorescence microscopy
BODIPY 558/568 C12 (RedC12, Life Technologies) was employed to measure β-oxidation as published previously (39). The cells were seeded at a density of 125 cells/mm2 in a 96-well CellCarrier plate (PerkinElmer Life Sciences). After adhering, the cells were maintained in RPMI medium supplemented with 5% FBS and 1 μm RedC12 for 16 h. Subsequently, the RPMI was replaced with complete medium containing 1% FBS and 50 μm etomoxir (or 0.1% DMSO) for 3 h. Alternatively, shGFP and shGLS cells without treatment were used. After that, mitochondria were labeled with 100 nm MitoTracker Deep Red (Life Technologies) and 2.5 μm Hoechst (Life Technologies, H3570) for 30 min. Images were then taken using an Operetta microscope (PerkinElmer Life Sciences). The analysis was performed with Columbus software (Perkin Elmer Life Sciences). The fluorescence intensity was reported as the mean value per cell. For colocalization, we calculated the overlap of RedC12 and MitoTracker Deep Red. Images were captured in Nipkow spinning disk confocal mode, and the analysis was performed with ImageJ software using the plugin Coloc2. Approximately 150 cells of each well were used to calculate Pearson's correlation coefficient (71).
Migration assay
In the wound-healing migration assays, 1875 cells/mm2 (HCC1806 and HCC70) or 937.5 cells/mm2 (BT549 and HCC1937) were seeded over 96-well plates pre-coated for 1 h with 300 μg/ml collagen type I from rat tails in acetic acid at 37 °C. After cell attachment for 16 h and serum starvation for 24 h, wounds were created with pipette tips, and the cells were immediately treated with 1 μm CB-839 and 50 μm etomoxir individually or in combination (DMSO vehicle at 0.2% (v/v)) in RPMI medium supplemented with 10% FBS. The cells were imaged for 18 h using Operetta (PerkinElmer Life Sciences) in bright-field mode every hour in a 5% CO2 atmosphere. The images were processed with Fiji-ImageJ using a macro based on previous work (72).
Lipid droplets and lysosomes staining
On a 96-well plate, 156.25 cells/mm2 were grown for 48 h in complete RPMI medium supplemented with 10% FBS. After medium removal, the cells were incubated with 500 nm LysoTracker Red DND-99 (Life Technologies) for 1 h at 37 °C and 5% CO2. The cells were rinsed with PBS and fixed with 3.7% formaldehyde in PBS added to 2.5 μm Hoechst for nucleus staining. For lipid labeling, the cells were incubated with 1:1000 LipidTOX neutral lipid (Life Technologies), and the plate was sealed with adherent film. Images were acquired immediately with an Operetta fluorescence microscope in Nipkow spinning disk confocal mode (19 stacks of 1-μm increments per field were collected). The intensity of lipid droplet labeling within the lysosomes (defined as regions of interest) was quantified using Harmony software. Representative images were obtained in the biological imaging facility of LNBio using a Leica TCS SP8 confocal mounted on a Leica DMI 6000 inverted microscope.
Transmission EM
A cell monolayer grown over a glass coverslip was fixed with 2.5% glutaraldehyde and 3 mm CaCl2 in 0.1 m sodium cacodylate buffer for 5 min at room temperature, followed by 1 h of incubation on ice. For lipid visualization using EM, imidazole-buffered osmium tetroxide was used as a stain as described previously (73). After fixation, the samples were washed three times in 0.1 m sodium cacodylate and 3 mm CaCl2 solution and post-fixed with 2% osmium tetroxide in 0.1 m imidazole buffer for 30 min and stained en bloc in ice-cold 2% uranyl acetate overnight. The cells were dehydrated in ethanol on ice, ending with four changes of 100% ethanol at room temperature. The dehydrated cells were infiltrated in Epon resin. After four changes of resin solution, a fifth resin change was performed, and the dish was immediately placed in a laboratory oven at 60 °C to be polymerized for 72 h. Ultrathin sections were cut with a Leica Ultracut microtome, stained with 2% uranyl acetate and Reynold's lead citrate, and then examined in an LEO 906-Zeiss transmission electron microscope (at the Electron Microscopy Laboratory of Institute of Biology, Campinas State University) using an accelerating voltage of 60 kV.
Author contributions
L. M. d. R. and S. M. G. D. conceptualization; L. M. d. R., D. A., and S. M. G. D. data curation; L. M. d. R. and S. M. G. D. formal analysis; L. M. d. R. and S. M. G. D. validation; L. M. d. R., D. A., R. O. O. S., C. F. R. A., K. R. S. d. O., F. C.-d.-S., F. M. d. S. P., M. M. D., and S. M. G. D. investigation; L. M. d. R. and S. M. G. D. visualization; L. M. d. R., D. A., R. O. O. S., F. C.-d.-S., M. M. D., and S. M. G. D. methodology; L. M. d. R. and S. M. G. D. writing-original draft; L. M. d. R., D. A., R. O. O. S., C. F. R. A., K. R. S. d. O., F. C.-d.-S., F. M. d. S. P., M. M. D., S. R. C., P. M. M. d. M.-V., A. M. S., and S. M. G. D. writing-review and editing; D. A. software; S. R. C., P. M. M. d. M.-V., A. M. S., and S. M. G. D. supervision; S. M. G. D. project administration.
Supplementary Material
Acknowledgments
We thank LNBio for access to core facilities. We also thank Dr. Alessandra Girasole for expert technical support and the TCGA Research Network.
This work was supported by São Paulo Research Foundation (FAPESP) fellowships to L. M. R. (2014/18061-9), D. A. (2014/17820-3), C. F. R. A. (2013/23510-4), F. C. S. (2017/06225-5), K. R. S. O. (2014/06512-6), and F. M. S. P. (2015/26059-7) and research grants to S. M. G. D. (2014/15968-3 and 2015/25832-4), A. M. S. (2016/06034-2), and P. M. M. d. M.-V. (2015/15626-8). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- TNBC
- triple-negative breast cancer
- GLS
- glutaminase
- GAC
- glutaminase C
- TCGA
- the cancer genome atlas
- AMPK
- AMP-activated protein kinase
- ACC
- acetyl-CoA carboxylase
- SER
- Saddles, Edges, Ridges
- DE
- differentially expressed
- FC
- fold change
- GO
- Gene Ontology
- OCR
- oxygen consumption rate
- FA
- fatty acid
- FPKM-UQ
- fragments per kilobase of transcript per million mapped reads upper quartile.
References
- 1. Prat A., and Perou C. M. (2011) Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 5, 5–23 10.1016/j.molonc.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernardi R., and Gianni L. (2014) Hallmarks of triple negative breast cancer emerging at last? Cell Res. 24, 904–905 10.1038/cr.2014.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaplan H. G., and Malmgren J. A. (2008) Impact of triple negative phenotype on breast cancer prognosis. Breast J. 14, 456–463 10.1111/j.1524-4741.2008.00622.x [DOI] [PubMed] [Google Scholar]
- 4. Turner N. C., and Reis-Filho J. S. (2013) Tackling the diversity of triple-negative breast cancer. Clin. Cancer Res. 19, 6380–6388 10.1158/1078-0432.CCR-13-0915 [DOI] [PubMed] [Google Scholar]
- 5. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 6. Pavlova N. N., and Thompson C. B. (2016) The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee K.-H., Hsu E.-C., Guh J.-H., Yang H.-C., Wang D., Kulp S. K., Shapiro C. L., and Chen C.-S. (2011) Targeting energy metabolic and oncogenic signaling pathways in triple-negative breast cancer by a novel adenosine monophosphate-activated protein kinase (AMPK) activator. J. Biol. Chem. 286, 39247–39258 10.1074/jbc.M111.264598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Noh S., Kim D. H., Jung W. H., and Koo J. S. (2014) Expression levels of serine/glycine metabolism-related proteins in triple negative breast cancer tissues. Tumour Biol. 35, 4457–4468 10.1007/s13277-013-1588-z [DOI] [PubMed] [Google Scholar]
- 9. Cao M. D., Lamichhane S., Lundgren S., Bofin A., Fjøsne H., Giskeødegård G. F., and Bathen T. F. (2014) Metabolic characterization of triple negative breast cancer. BMC Cancer 14, 941 10.1186/1471-2407-14-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lim S.-O., Li C.-W., Xia W., Lee H.-H., Chang S.-S., Shen J., Hsu J. L., Raftery D., Djukovic D., Gu H., Chang W.-C., Wang H.-L., Chen M.-L., Huo L., Chen C.-H., et al. (2016) EGFR signaling enhances aerobic glycolysis in triple-negative breast cancer cells to promote tumor growth and immune escape. Cancer Res. 76, 1284–1296 10.1158/1538-7445.AM2016-1284,10.1158/0008-5472.CAN-15-2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen L., O'Shea J. M., Kaadige M. R., Cunha S., Wilde B. R., Cohen A. L., Welm A. L., and Ayer D. E. (2015) Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc. Natl. Acad. Sci. 112, 5425–5430 10.1073/pnas.1501555112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Toole S. A., Beith J. M., Millar E. K., West R., McLean A., Cazet A., Swarbrick A., and Oakes S. R. (2013) Therapeutic targets in triple negative breast cancer. J. Clin. Pathol. 66, 530–542 10.1136/jclinpath-2012-201361 [DOI] [PubMed] [Google Scholar]
- 13. Farabegoli F., Vettraino M., Manerba M., Fiume L., Roberti M., and Di Stefano G. (2012) Galloflavin, a new lactate dehydrogenase inhibitor, induces the death of human breast cancer cells with different glycolytic attitude by affecting distinct signaling pathways. Eur. J. Pharm. Sci. 47, 729–738 10.1016/j.ejps.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 14. Timmerman L. A., Holton T., Yuneva M., Louie R. J., Padró M., Daemen A., Hu M., Chan D. A., Ethier S. P., van 't Veer L. J., Polyak K., McCormick F., and Gray J. W. (2013) Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24, 450–465 10.1016/j.ccr.2013.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Geldermalsen M., Wang Q., Nagarajah R., Marshall A. D., Thoeng A., Gao D., Ritchie W., Feng Y., Bailey C. G., Deng N., Harvey K., Beith J. M., Selinger C. I., O'Toole S. A., Rasko J. E., and Holst J. (2016) ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene 35, 3201–3208 10.1038/onc.2015.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang L., Moss T., Mangala L. S., Marini J., Zhao H., Wahlig S., Armaiz-Pena G., Jiang D., Achreja A., Win J., Roopaimoole R., Rodriguez-Aguayo C., Mercado-Uribe I., Lopez-Berestein G., Liu J., et al. (2014) Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol. Syst. Biol. 10, 728–728 10.1002/msb.20134892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seltzer M. J., Bennett B. D., Joshi A. D., Gao P., Thomas A. G., Ferraris D. V., Tsukamoto T., Rojas C. J., Slusher B. S., Rabinowitz J. D., Dang C. V., and Riggins G. J. (2010) Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 70, 8981–8987 10.1158/0008-5472.CAN-10-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moncada S., Higgs E. A., and Colombo S. L. (2012) Fulfilling the metabolic requirements for cell proliferation. Biochem. J. 446, 1–7 10.1042/BJ20120427 [DOI] [PubMed] [Google Scholar]
- 19. Son J., Lyssiotis C. A., Ying H., Wang X., Hua S., Ligorio M., Perera R. M., Ferrone C. R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J. B., Bardeesy N., Asara J. M., Haigis M. C., et al. (2013) Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 10.1038/nature12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dang C. V. (2010) Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle 9, 3884–3886 10.4161/cc.9.19.13302 [DOI] [PubMed] [Google Scholar]
- 21. Ryu J. M., Lee S. H., Seong J. K., and Han H. J. (2015) Glutamine contributes to maintenance of mouse embryonic stem cell self-renewal through PKC-dependent downregulation of HDAC1 and DNMT1/3a. Cell Cycle 14, 3292–3305 10.1080/15384101.2015.1087620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun H.-W., Yu X.-J., Wu W.-C., Chen J., Shi M., Zheng L., and Xu J. (2016) GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PLoS ONE 11, e0168907 10.1371/journal.pone.0168907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Márquez J., López de la Oliva A. R., Matés J. M., and Segura J. A., and Alonso F. J. (2006) Glutaminase: a multifaceted protein not only involved in generating glutamate. Neurochem. Int. 48, 465–471 10.1016/j.neuint.2005.10.015 [DOI] [PubMed] [Google Scholar]
- 24. Pérez-Gómez C., Matés J. M., Gómez-Fabre P. M., del Castillo-Olivares A., Alonso F. J., and Márquez J. (2003) Genomic organization and transcriptional analysis of the human l-glutaminase gene. Biochem. J. 370, 771–784 10.1042/bj20021445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson M. M., McBryant S. J., Tsukamoto T., Rojas C., Ferraris D. V., Hamilton S. K., Hansen J. C., and Curthoys N. P. (2007) Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 406, 407–414 10.1042/BJ20070039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Q., Stalnecker C., Zhang C., McDermott L. A., Iyer P., O'Neill J., Reimer S., Cerione R. A., and Katt W. P. (2018) Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism. J. Biol. Chem. 293, 3535–3545 10.1074/jbc.M117.810101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song M., Kim S.-H., Im C. Y., and Hwang H.-J. (2018) Recent development of small molecule glutaminase inhibitors. Curr. Top. Med. Chem. 18, 432–443 10.2174/1568026618666180525100830 [DOI] [PubMed] [Google Scholar]
- 28. Gross M. I., Demo S. D., Dennison J. B., Chen L., Chernov-Rogan T., Goyal B., Janes J. R., Laidig G. J., Lewis E. R., Li J., Mackinnon A. L., Parlati F., Rodriguez M. L., Shwonek P. J., Sjogren E. B., et al. (2014) Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 10.1158/1535-7163.MCT-13-0870 [DOI] [PubMed] [Google Scholar]
- 29. Garber K. (2016) Cancer anabolic metabolism inhibitors move into clinic. Nat. Biotechnol. 34, 794–795 10.1038/nbt0816-794 [DOI] [PubMed] [Google Scholar]
- 30. Liu K., and Czaja M. J. (2013) Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20, 3–11 10.1038/cdd.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park S. H., Gammon S. R., Knippers J. D., Paulsen S. R., Rubink D. S., and Winder W. W. (2002) Phosphorylation-activity relationships of AMPK and acetyl-CoA carboxylase in muscle. J. Appl. Physiol. 92, 2475–2482 10.1152/japplphysiol.00071.2002 [DOI] [PubMed] [Google Scholar]
- 32. Camarda R., Zhou A. Y., Kohnz R. A., Balakrishnan S., Mahieu C., Anderton B., Eyob H., Kajimura S., Tward A., Krings G., Nomura D. K., and Goga A. (2016) Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 22, 427–432 10.1038/nm.4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park J. H., Vithayathil S., Kumar S., Sung P.-L., Dobrolecki L. E., Putluri V., Bhat V. B., Bhowmik S. K., Gupta V., Arora K., Wu D., Tsouko E., Zhang Y., Maity S., Donti T. R., et al. (2016) Fatty acid oxidation-driven Src links mitochondrial energy reprogramming and oncogenic properties in triple-negative breast cancer. Cell Rep. 14, 2154–2165 10.1016/j.celrep.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blomme A., Costanza B., de Tullio P., Thiry M., Van Simaeys G., Boutry S., Doumont G., Di Valentin E., Hirano T., Yokobori T., Gofflot S., Peulen O., Bellahcène A., Sherer F., Le Goff C., et al. (2017) Myoferlin regulates cellular lipid metabolism and promotes metastases in triple-negative breast cancer. Oncogene 36, 2116–2130 10.1038/onc.2016.369 [DOI] [PubMed] [Google Scholar]
- 35. Wright H. J., Hou J., Xu B., Cortez M., Potma E. O., Tromberg B. J., and Razorenova O. V. (2017) CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. Proc. Natl. Acad. Sci. 114, E6556–E6565 10.1073/pnas.1703791114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Bliek A. M., Shen Q., and Kawajiri S. (2013) Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chris Bakal and Dr Julia Sero. (2012) High content image analysis phenotypic characterization of mitochondria in breast cancer cells using morphology and texture properties [Google Scholar]
- 38. Quintero M., Adamoski D., Reis L. M. D., Ascenção C. F. R., Oliveira K. R. S., Gonçalves K. A., Dias M. M., Carazzolle M. F., and Dias S. M. G. (2017) Guanylate-binding protein-1 is a potential new therapeutic target for triple-negative breast cancer. BMC Cancer 17, 727 10.1186/s12885-017-3726-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rambold A. S., Cohen S., and Lippincott-Schwartz J. (2015) Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32, 678–692 10.1016/j.devcel.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lopaschuk G. D., McNeil G. F., and McVeigh J. J. (1989) Glucose oxidation is stimulated in reperfused ischemic hearts with the carnitine palmitoyltransferase 1 inhibitor, etomoxir. Mol. Cell. Biochem. 88, 175–179 [DOI] [PubMed] [Google Scholar]
- 41. Rodrigues M. F., Obre E., de Melo F. H., Santos G. C. Jr., Galina A., Jasiulionis M. G., Rossignol R., Rumjanek F. D., and Amoêdo N. D. (2016) Enhanced OXPHOS, glutaminolysis and -oxidation constitute the metastatic phenotype of melanoma cells. Biochem. J. 473, 703–715 10.1042/BJ20150645 [DOI] [PubMed] [Google Scholar]
- 42. Merrill G. F., Kurth E. J., Hardie D. G., and Winder W. W. (1997) AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am. J. Physiol. 273, E1107–12 [DOI] [PubMed] [Google Scholar]
- 43. Momcilovic M., Bailey S. T., Lee J. T., Fishbein M. C., Magyar C., Braas D., Graeber T., Jackson N. J., Czernin J., Emberley E., Gross M., Janes J., Mackinnon A., Pan A., Rodriguez M., et al. (2017) Targeted Inhibition of EGFR and glutaminase induces metabolic crisis in EGFR mutant lung cancer. Cell Rep. 18, 601–610 10.1016/j.celrep.2016.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., and Moller D. E. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 10.1172/JCI13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lampa M., Arlt H., He T., Ospina B., Reeves J., Zhang B., Murtie J., Deng G., Barberis C., Hoffmann D., Cheng H., Pollard J., Winter C., Richon V., Garcia-Escheverria C., et al. (2017) Glutaminase is essential for the growth of triple-negative breast cancer cells with a deregulated glutamine metabolism pathway and its suppression synergizes with mTOR inhibition. PLoS ONE 12, e0185092 10.1371/journal.pone.0185092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dornier E., Rabas N., Mitchell L., Novo D., Dhayade S., Marco S., Mackay G., Sumpton D., Pallares M., Nixon C., Blyth K., Macpherson I. R., Rainero E., and Norman J. C. (2017) Glutaminolysis drives membrane trafficking to promote invasiveness of breast cancer cells. Nat. Commun. 8, 2255 10.1038/s41467-017-02101-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peyton K. J., Liu X. M., Yu Y., Yates B., Behnammanesh G., and Durante W. (2018) Glutaminase-1 stimulates the proliferation, migration, and survival of human endothelial cells. Biochem. Pharmacol. 156, 204–214 10.1016/j.bcp.2018.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ascenção C. F. R., Nagampalli R. S. K., Islam Z., Pinheiro M. P., Menezes Dos Reis L., Pauletti B. A., de Guzzi Cassago C. A., Granato D. C., Paes Leme A. F., and Dias S. M. G. (2018) N-terminal phosphorylation of glutaminase C decreases its enzymatic activity and cancer cell migration. Biochimie 154, 69–76 10.1016/j.biochi.2018.07.022 [DOI] [PubMed] [Google Scholar]
- 49. Chakrabarti G., Moore Z. R., Luo X., Ilcheva M., Ali A., Padanad M., Zhou Y., Xie Y., Burma S., Scaglioni P. P., Cantley L. C., DeBerardinis R. J., Kimmelman A. C., Lyssiotis C. A., and Boothman D. A. (2015) Targeting glutamine metabolism sensitizes pancreatic cancer to PARP-driven metabolic catastrophe induced by β-lapachone. Cancer Metab. 3, 12 10.1186/s40170-015-0137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jacque N., Ronchetti A. M., Larrue C., Meunier G., Birsen R., Willems L., Saland E., Decroocq J., Maciel T. T., Lambert M., Poulain L., Hospital M. A., Sujobert P., Joseph L., Chapuis N., et al. (2015) Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 126, 1346–1356 10.1182/blood-2015-01-621870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kalinsky K., Harding J., DeMichele A., Infante J., Gogineni K., Owonikoko T., Isakoff S., Iliopoulos O., Patel M., Munster P., Telli M., Jenkins Y., Fiji G., Whiting S., and Meric-Bernstam F. (2018) Abstract PD3–13: phase 1 study of CB-839, a first-in-class oral inhibitor of glutaminase, in combination with paclitaxel in patients with advanced triple negative breast cancer. Cancer Res. 78, PD3–13 10.1158/1538-7445.SABCS17-PD3-13 [DOI] [Google Scholar]
- 52. Cluntun A. A., Lukey M. J., Cerione R. A., and Locasale J. W. (2017) Glutamine metabolism in cancer: understanding the heterogeneity. Trends in Cancer 3, 169–180 10.1016/j.trecan.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hermanova I., Arruabarrena-Aristorena A., Valis K., Nuskova H., Alberich-Jorda M., Fiser K., Fernandez-Ruiz S., Kavan D., Pecinova A., Niso-Santano M., Zaliova M., Novak P., Houstek J., Mracek T., Kroemer G., et al. (2016) Pharmacological inhibition of fatty-acid oxidation synergistically enhances the effect of l-asparaginase in childhood ALL cells. Leukemia 30, 209–218 10.1038/leu.2015.213 [DOI] [PubMed] [Google Scholar]
- 54. Zaugg K., Yao Y., Reilly P. T., Kannan K., Kiarash R., Mason J., Huang P., Sawyer S. K., Fuerth B., Faubert B., Kalliomäki T., Elia A., Luo X., Nadeem V., Bungard D., et al. (2011) Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 25, 1041–1051 10.1101/gad.1987211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Long B., Muhamad R., Yan G., Yu J., Fan Q., Wang Z., Li X., Purnomoadi A., Achmadi J., and Yan X. (2016) Quantitative proteomics analysis reveals glutamine deprivation activates fatty acid β-oxidation pathway in HepG2 cells. Amino Acids. 48, 1297–1307 10.1007/s00726-016-2182-7 [DOI] [PubMed] [Google Scholar]
- 56. Halama A., Kulinski M., Dib S. S., Zaghlool S. B., Siveen K. S., Iskandarani A., Zierer J., Prabhu K. S., Satheesh N. J., Bhagwat A. M., Uddin S., Kastenmüller G., Elemento O., Gross S. S., and Suhre K. (2018) Accelerated lipid catabolism and autophagy are cancer survival mechanisms under inhibited glutaminolysis. Cancer Lett. 430, 133–147 10.1016/j.canlet.2018.05.017 [DOI] [PubMed] [Google Scholar]
- 57. Yao C.-H., Liu G.-Y., Wang R., Moon S. H., Gross R. W., and Patti G. J. (2018) Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation. PLoS Biol. 16, e2003782 10.1371/journal.pbio.2003782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., and Novina C. D. (2003) Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 10.1261/rna.2192803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bernt E., and Bergmeyer H. U. (1974) l-Glutamate UV-assay with glutamate dehydrogenase and NAD. in Methods of Enzymatic Analysis, 2nd Ed (Bergmeyer H. U., and Gawehn K. eds), pp. 1704–1715, Academic Press Inc., New York and London [Google Scholar]
- 60. Cassago A., Ferreira A. P., Ferreira I. M., Fornezari C., Gomes E. R., Greene K. S., Pereira H. M., Garratt R. C., Dias S. M., and Ambrosio A. L. (2012) Mitochondrial localization and structure-based phosphate activation mechanism of glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. 109, 1092–1097 10.1073/pnas.1112495109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kenny J., Bao Y., Hamm B., Taylor L., Toth A., Wagers B., and Curthoys N. P. (2003) Bacterial expression, purification, and characterization of rat kidney-type mitochondrial glutaminase. Protein Expr. Purif. 31, 140–148 10.1016/S1046-5928(03)00161-X [DOI] [PubMed] [Google Scholar]
- 62. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 63. Daemen A., Griffith O. L., Heiser L. M., Wang N. J., Enache O. M., Sanborn Z., Pepin F., Durinck S., Korkola J. E., Griffith M., Hur J. S., Huh N., Chung J., Cope L., Fackler M. J., et al. (2013) Modeling precision treatment of breast cancer. Genome Biol. 14, R110 10.1186/gb-2013-14-10-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Varley K. E., Gertz J., Roberts B. S., Davis N. S., Bowling K. M., Kirby M. K., Nesmith A. S., Oliver P. G., Grizzle W. E., Forero A., Buchsbaum D. J., LoBuglio A. F., and Myers R. M. (2014) Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res. Treat. 146, 287–297 10.1007/s10549-014-3019-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Young M. D., Wakefield M. J., Smyth G. K., and Oshlack A. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li J., Lu Y., Akbani R., Ju Z., Roebuck P. L., Liu W., Yang J.-Y., Broom B. M., Verhaak R. G. W., Kane D. W., Wakefield C., Weinstein J. N., Mills G. B., and Liang H. (2013) TCPA: a resource for cancer functional proteomics data. Nat. Methods. 10, 1046–1047 10.1038/nmeth.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Karlic H., Lohninger S., Koeck T., and Lohninger A. (2002) Dietary L-carnitine stimulates carnitine acyltransferases in the liver of aged rats. J. Histochem. Cytochem. 50, 205–212 10.1177/002215540205000208 [DOI] [PubMed] [Google Scholar]
- 70. Huynh F. K., Green M. F., Koves T. R., and Hirschey M. D. (2014) Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 542, 391–405 10.1016/B978-0-12-416618-9.00020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Adler J., and Parmryd I. (2010) Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A 77, 733–742 [DOI] [PubMed] [Google Scholar]
- 72. Silva Nunes J. P., and Martins Dias A. A. (2017) ImageJ macros for the user-friendly analysis of soft-agar and wound-healing assays. BioTechniques 10.2144/000114535 [DOI] [PubMed] [Google Scholar]
- 73. Angermüller S., and Fahimi H. D. (1982) Imidazole-buffered osmium tetroxide: an excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 14, 823–835 10.1007/BF01033631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.