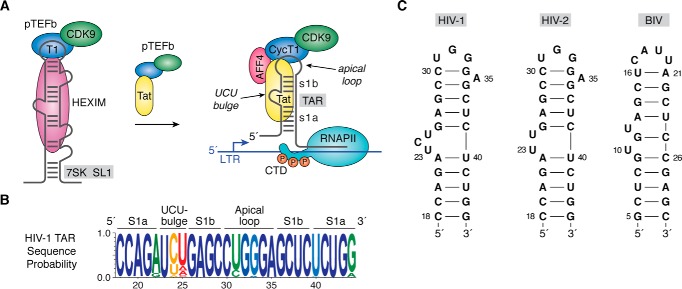
Figure 1.
HIV-1 TAR role in transcription, sequence conservation, and secondary structure. A, cartoon diagram of an inactive pTEFb complex comprising CDK9 and CycT1 in the context of HEXIM protein bound to 7SK ncRNA in the host. The arrow indicates that addition of the HIV-1 regulatory protein Tat competes with HEXIM, removing pTEFb from 7SK, which is then escorted by Tat to the TAR RNA element of HIV-1 (141). TAR is essential for transcription and is depicted as a stem loop interrupted by a central bulge that comprises nucleotides 18–44 of the viral transcript (78, 142). Tat interacts directly with TAR and promotes formation of a host SEC comprising pTEFb, scaffold proteins such as AFF4, and other factors (43, 47, 52, 68, 143, 144). CDK9 phosphorylates host RNA polymerase II in its CTD, which releases pausing and stimulates synthesis of full-length viral transcripts (33, 145–147). B, Web-logo showing the sequence conservation of HIV-1 TAR based on circulating forms of the virus compiled as described (77); blue represents the greatest conservation, and red indicates the poor conservation. Elements of the secondary structure including helical stems s1a and s1b are labeled. C, secondary structures of various TAR RNAs. The canonical Cyt30–Gua34 pair of HIV-1 TAR is supported by chemical modification, NMR, sequence conservation, and CycT1-binding requirements (65, 66, 77, 148, 149). A key difference between HIV-1 and HIV-2 TAR is deletion of Cyt24 in the central bulge (150). Details of the BIV TAR secondary structure were derived from Refs. 73, 75, 151. CTD, C-terminal domain.