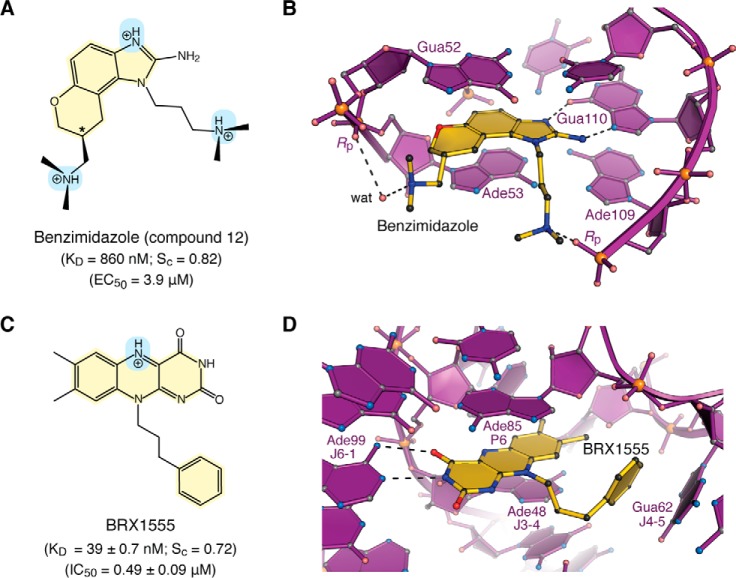
Figure 4.
Chemical properties and modes of drug binding to representative ncRNAs. A, chemical structure of benzimidazole variant “compound 12”. The KD of binding to HCV domain IIa and EC50 from replicon assays are shown (118). The chiral center is labeled with an asterisk. B, ball–and–stick diagram of HCV domain IIa (purple) bound to compound 12 (yellow) (PDB entry 3tzr) (28). The stereochemistry was not resolved in electron density maps. C, chemical structure of the FMN analogue BRX1555. The KD value of drug binding, the EC50 value from single-round transcription assays, and the IC50 value for bacterial growth inhibition are provided (25). D, ball-and-stick diagram of the Fusobacterium nucleatum FMN riboswitch in complex with BRX1555 (PDB entry 6dn3) (25). The respective ncRNA–inhibitor structures were chosen based on visual inspection of ligand fit to electron-density maps and associated quality-control indicators.