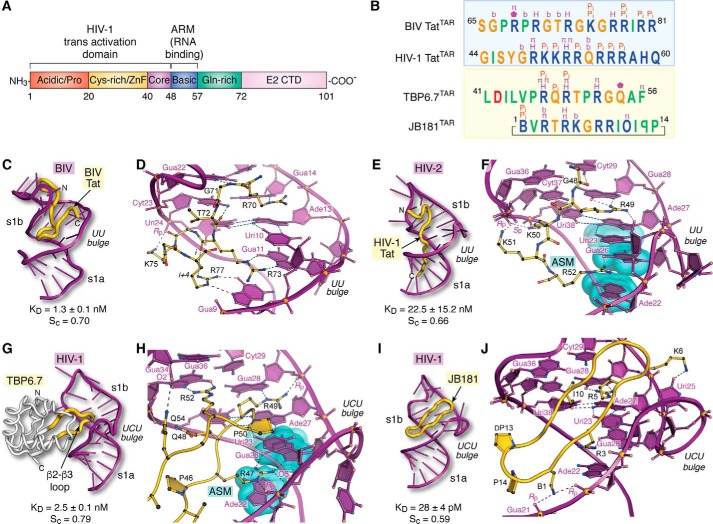
Figure 5.
Tat organization and molecular recognition of TAR by Tat ARM domains, lab-evolved proteins, and cyclic peptides. A, functional domain organization of the HIV-1 Tat protein (154). The transactivation domain comprises an N-terminal acidic and proline-rich domain, a cysteine-rich domain that binds Zn(II) (i.e. zinc finger or ZnF), a core region, and a basic ARM. Additional domains include a glutamine-rich region and the E2 CTD. B, summary of peptide sequence interactions with TAR RNAs from C–J (blue box). Amino acids of naturally occurring BIV and HIV-1 Tat ARMs are shown; the sequence alignment is based on common recognition modes of RNA targets as follows: Gua11(26) and Gua14(28) of BIV(HIV) TAR are recognized by Arg-73(R52) and Arg-70(R49) of BIV(HIV) Tat (yellow box). Amino acids of lab-evolved proteins and cyclic peptides from structure-based design. The sequence alignment is based on common spatial recognition at Gua26 and Gua28 of HIV-1 TAR by Arg-47 and Arg-49 of TBP6.7 and Arg-3 and Arg-5 of JB181. Specific RNA–peptide interaction types are listed above each amino acid; the symbols are as follows: π indicates cation–π or aromatic stacking; H equals hydrogen-bond recognition of a guanine Hoogsteen edge; Pi indicates salt-bridge formation to the phosphate backbone; b indicates hydrogen-bond recognition to a nucleobase; a pentagon indicates a hydrogen-bond contact to ribose. Symbols of nonstandard amino acids are as follows: B equals l-2,4-diaminobutyric acid; O equals l-ornithine; backward P is d-proline. C, global view depicting BIV TAR (purple ribbon) recognition by the BIV-Tat ARM (yellow worm) (PDB entry 1biv) (75). The Sc value was calculated from the lowest energy NMR core structure (amino acids 67–79). D, close-up view of BIV TAR recognition by BIV Tat at the UU bulge. Despite differences in the central bulge compared with HIV TAR (Fig. 1C), BIV TAR exhibits a major-groove base triple at Uri10·Ade13–Uri24. The Tat peptide undergoes a sharp bend with dihedral angles of ϕi + 1 75°, ψi + 1 −10°, and ϕi + 2 −133°, ψi + 2 63° characteristic of a type V′ turn (130); the peptide has no other β-hairpin characteristics. For clarity, only amino acids engaged in peptide–RNA were included in diagrams. E, global view depicting HIV-2 TAR recognition by the HIV-1 Tat peptide (PDB entry 6mce) (76). The Sc value was calculated from the lowest energy NMR core structure (amino acids 48–54). F, close-up view of HIV-2 TAR recognition at its UU bulge by HIV-1 Tat. Three nucleobases compose an ASM (cyan highlight) that engages Arg-52 of Tat. G, global view depicting HIV-1 TAR recognition by TBP6.7 (PDB entry 6cmn) (77). The lab-evolved β2–β3 loop (yellow) recognizes the TAR major groove. The Sc value was calculated from the co-crystal structure. H, close-up view of HIV-1 TAR recognition in the central UCU bulge by TBP6.7. Arg-47 engages TAR at the ASM, similar to F. I, global view of HIV-1 TAR recognition by the cyclic peptide JB181 (PDB entry 6d2u) (63). The Sc value was calculated from the lowest energy NMR structure. J, close-up view of HIV-1 TAR recognition at the UCU bulge by JB181. The DP13–LP14 turn is shown to emphasize the restrained peptide conformation. CTD, C-terminal domain.