Figure 6.
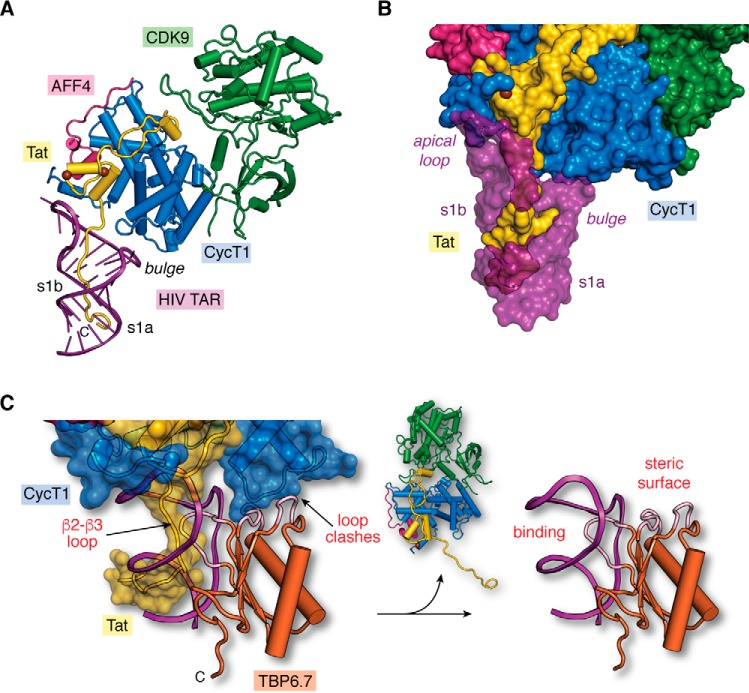
Structural model of the core super-elongation complex bound to HIV TAR(1–60). A, hypothetical model of HIV-1 TAR in complex with CycT1–CDK9–AFF4–Tat(1–60). The putative Tat ARM trajectory is based on superposition of HIV-1 TAR in the context of the recent co-crystal structure of the SEC core complex (PDB entry 6cyt) (74) upon HIV-2 TAR in the context of the recent TAR–Tat(44–60) structure (PDB entry 6mce) (76), as depicted in Fig. 5C. Rather than localizing the Tat ARM domain to the s1b stem (74), the model predicts that the ARM runs through TAR's major groove. B, surface model of the hypothetical SEC–TAR–Tat model from A. The surface emphasizes three virus–host protein contact points to TAR. First, the RNA apical loop makes a modest number of interactions with Tat (yellow) and CycT1 (blue). Second, the proximal segment of the Tat ARM (amino acids 47–52) contacts TAR within s1b and the bulged loop. Third, the distal Tat ARM (amino acids 53–57) contacts TAR at s1a. For emphasis, TAR is depicted as a semi-transparent surface to allow Tat visualization in the major groove. Notably, there are no observed contacts between the TAR bulge and CycT1, which is behind the RNA in this orientation. C, two sites of steric blocking are predicted when TBP6.7 is docked onto the SEC–TAR–Tat model of A. Specifically, interference occurs by the β2–β3 loop of TBP6.7 (labeled “binding”) where the Tat ΑRΜ interacts with s1b and the UCU bulge of TAR (Fig. 5, G and H). TBP6.7 loops also interfere with the positioning of CycT1 in the context of the SEC due to steric clashes (labeled “steric surface”). The net result is displacement of the SEC, resulting in the TAR–TBP6.7 complex (right panel). The hypothetical model (left panel) was prepared by superposition of the HIV-1 TAR–TBP6.7 co-crystal structure (PDB entry 6cmn) (77) upon the SEC–TAR–Tat model of A.