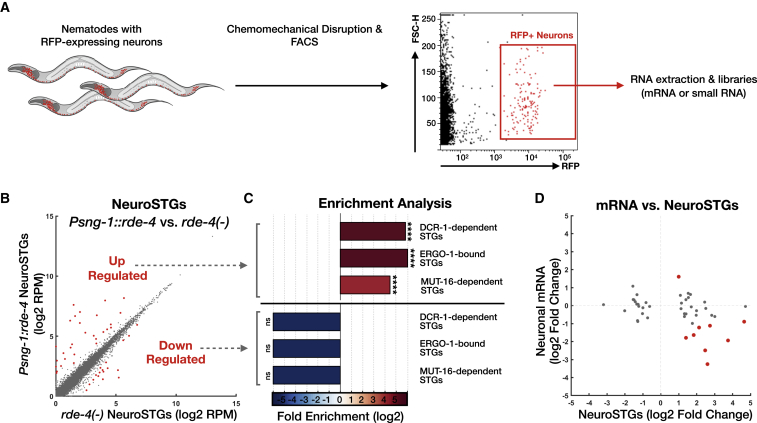
Figure 2.
Sorting of Neurons Followed by RNA Sequencing Allows the Characterization of Neuronal RDE-4-Dependent Small RNAs
(A) Scheme depicting the production of RNA libraries specifically from neurons of C. elegans. Single-cell suspensions were produced out of wild type, rde-4(−), and Psng-1::rde-4 strains that express Prab-3::rfp in neurons (Kaletsky et al., 2016, Stefanakis et al., 2015), followed by immediate fluorescence-activated cell sorting (FACS). Sorted RFP+ neurons were collected for total RNA isolation.
(B) Expression levels of NeuroSTGs in rescued Psng-1::rde-4 worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of rpm) of NeuroSTGs (see also Table S5). Each dot represents a NeuroSTG. 46 NeuroSTGs (red) displayed differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(C) x-fold enrichment or depletion values of upregulated NeuroSTGs (left) and downregulated NeuroSTGs (right) following neuronal RDE-4 rescue. See also Figure 1E. For the clarity of display, complete depletion (linear enrichment = 0) appears with the smallest value in the scale. ns, p > 0.05; ∗∗∗∗p < 10−4.
(D) Changes in neuronal mRNA levels (y axis) in Psng-1::rde-4 compared to rde-4(−) neurons, plotted against changes in their associated NeuroSTGs (x axis). Each dot represents the values for one gene, and the 46 genes with significant changes in their corresponding NeuroSTGs are shown (analyzed with Deseq2, adjusted p value < 0.1). Nine genes (red) exhibited also differential mRNA expression (analyzed with Deseq2, adjusted p value < 0.1).