Figure 3.
The Germline of Psng-1::rde-4 Worms Is Devoid of Functional RDE-4
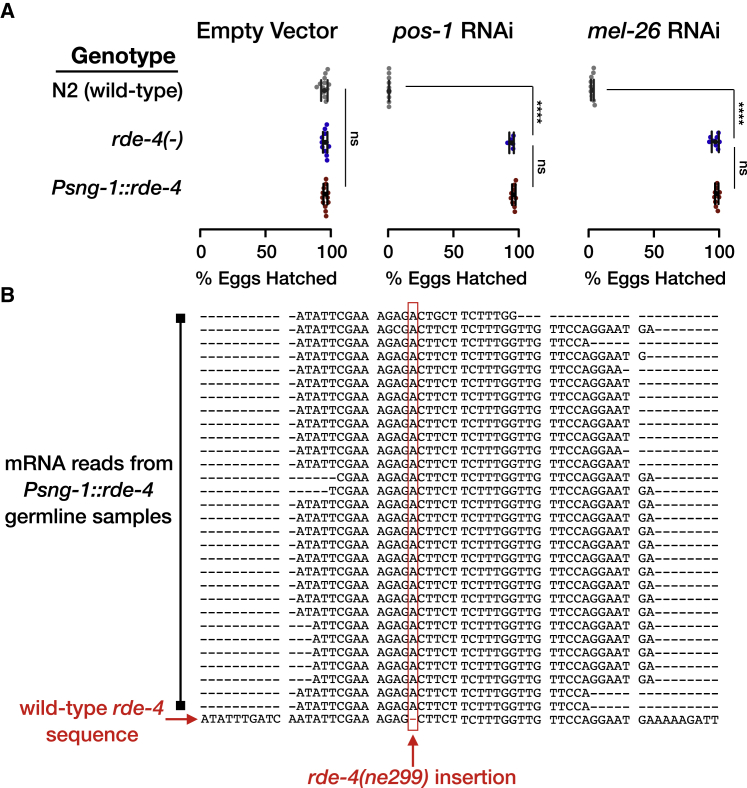
(A) Worms with the indicated genotype (y axis) were allowed to lay eggs on plates containing dsRNA-producing bacteria targeting the germline-expressed genes pos-1 and mel-26 or an empty-vector control. Shown are the percentage of hatched eggs per plate (x axis) following exposure to RNAi. Each dot represents one tested plate (biological replicate). Bars represent mean ± SD. Each group was tested in at least three independent experiments. p values were determined by two-way ANOVA with Tukey’s post hoc correction for multiple comparison, ∗∗∗∗p < 10−4; ns, p > 0.05.
(B) Multiple sequence alignment of all the sequencing reads aligned to the genomic locations in the vicinity of the insertion defining the rde-4(ne299) allele. We combined all the reads (30) obtained from three independent replicate gonads samples from Psng-1::rde-4 worms. The wild-type rde-4 sequence is shown at the bottom row. We display only reads in which the insertion site is neither in the edge of the read nor included in soft clipping region of the CIGAR string. Shown is the complementary strand of the rde-4 gene, with the insertion position (chr-III: 10,218,186) marked in a red rectangle.
See also Figure S3.