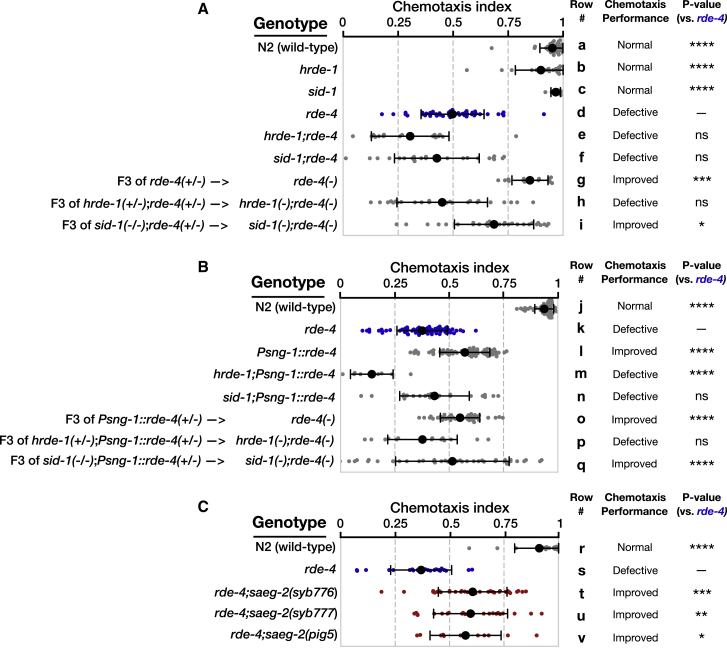
Figure 6.
Neuronal RDE-4 Controls Behavior Transgenerationally via the Germline Small RNA Machinery
Results for experiments testing chemotaxis to benzaldehyde (1:100) at day 1 of adulthood of worms (ethanol was used as control odor). Chemotaxis index = ((# worms at benzaldehyde) − (# worms at ethanol))/((# total worms on plate) − (# worms at origin)). Each dot represents one plate with >200 worms. All groups were tested on at least three independent trials, each including several biological replicates. Black bars represent mean ± SD. For convenience, each biological group was assigned a letter label. p values were determined by Kruskal-Wallis test with Dunn’s post hoc correction for multiple comparison to the rde-4(−) group. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 10−4.
(A) Chemotaxis experiments on the F3 rde-4(−) progeny of rde-4(+/−) P0 ancestors and their hrde-1 and sid-1 double mutants, together with control strains.
(B) Chemotaxis experiments on the F3 rde-4(−) progeny of Psng-1::rde-4(+/−);rde-4(−/−) P0 ancestors and their hrde-1 and sid-1 double mutants, together with control strains.
(C) Chemotaxis experiments on the rde-4;saeg-2 double mutants alleles, together with control strains. Three double mutant strains were generated via CRISPR/Cas9.
See also Figure S5.