Figure S1.
High-Copy Expression of RDE-4 in Neurons Regulates a Subset of STGs, Related to Figure 1
(A) A typical image demonstrating the neuronal expression pattern of the rescued RDE-4 (Prgef-1::rde-4::SL2::yfp), as monitored by examination of a trans-spliced YFP fluorescent reporter. Prgef-1::rde-4::SL2::yfp was co-injected with Punc122::GFP expressed in coelomocytes (marked in red). Bar = 20 μm.
(B and C) smFISH staining of yfp transcripts (magenta) and DAPI nuclei staining (blue) in one typical worm expressing Prgef-1::rde-4::SL2::yfp. Shown are focal plains focusing on the neuronal ventral chord (B), yellow dashed lines), and the germline (C), white dashed lines). Bar = 20 μm.
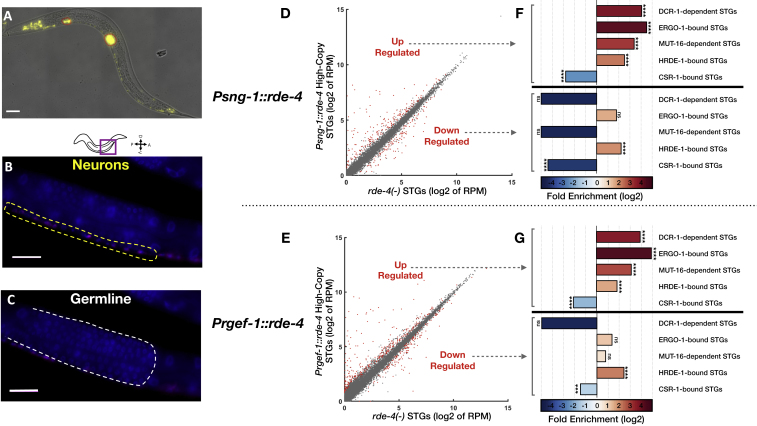
(D and E) Expression of STGs in rescued Psng-1::rde-4 (D) and Prgef-1::rde-4 (E) worms (y axis) compared to rde-4(ne299) mutants (x axis). Shown are the averaged expression values (log2 of RPM) of STGs (See also Table S2). Each dot represents an STG. Red dots: STGs that display differential expression between groups (analyzed with Deseq2, adjusted p value < 0.1).
(F and G) x-fold enrichment and depletion values of upregulated STGs (upper panel) and downregulated STGs (lower panel) following RDE-4 High-Copy rescue in neurons. P values for enrichment were calculated using 10,000 random gene sets identical in size to the tested group (See “STAR Methods” for details). For the clarity of display, complete depletion (linear enrichment = 0) appears with the smallest value in the scale. Enrichments were considered significant if p < 0.05. ns- p > 0.05. ∗∗∗- p < 0.001. ∗∗∗∗- p < 10−4.