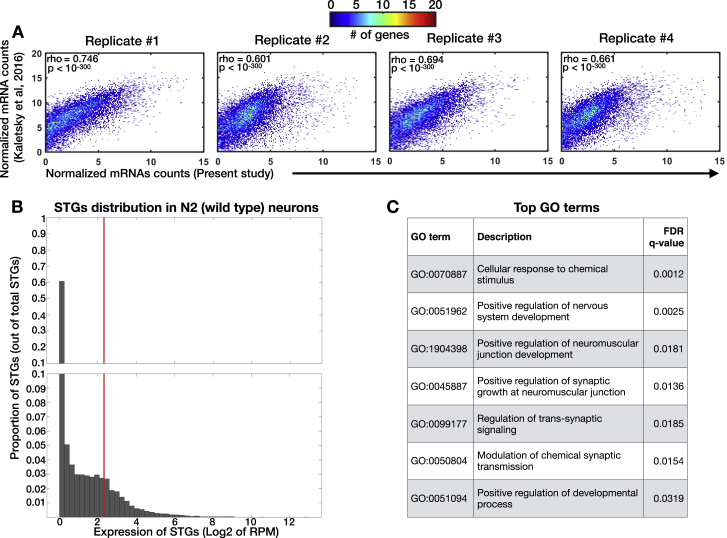
Figure S2.
Isolation and Sequencing of Neuronal Small RNAs and Neuronal mRNAs from N2 Wild-type Worms, Related to Figure 2
(A) Normalized mRNA levels reported by Kaletsky et al. (2016) (y axis), versus the mRNA levels (log2 of RPM) measured in four samples of N2 worms collected in independent experiments (x axis). Each dot represents a gene. As many points may overlap each other, and to better visualize the distribution of the data, we added a color code reflecting the number of genes in each bin.
(B) Shown is a histogram indicating the proportion of STGs (y axis) with different expression levels, displayed by log2 of RPM average (x axis). The vertical red line corresponds to value of 5 RPM (linear scale). We used a cut-off of > 5RPM to create a list of robustly expressed STGs in N2 neurons (See Table S3). Please note the scale in the y axis is changing, with steps of 0.01 in the range of 0 to 0.1, and steps of 0.1 for proportions higher than 0.1.
(C) Enriched GO terms for the sub-set of 412 genes targeted by NeuroSTGs with RPM > 5, which are upregulated in isolated neurons in comparison to STGs extracted from the entire animal. As can be seen, 6 of the 7 most enriched GO terms depict a variety of neuronal processes. Analysis done using the GOrilla tool (Eden et al., 2009); all the enriched GO terms with FDR < 0.05 are displayed.