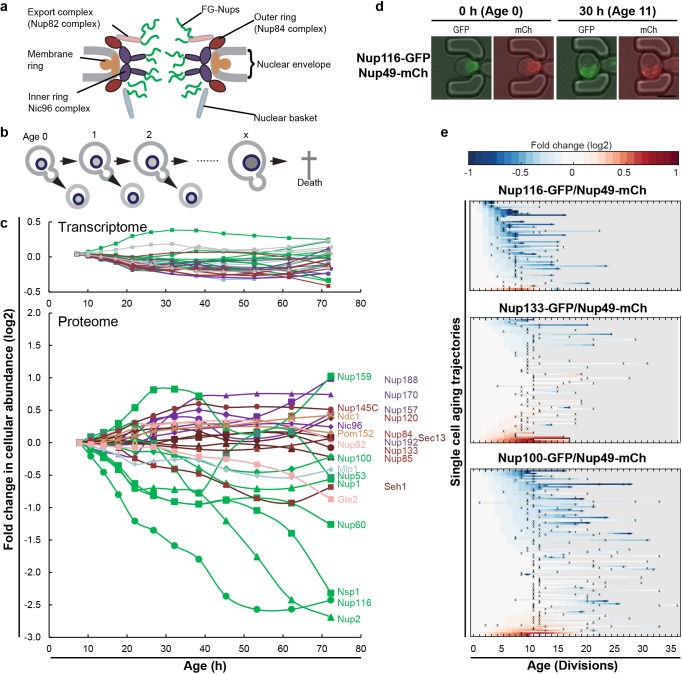
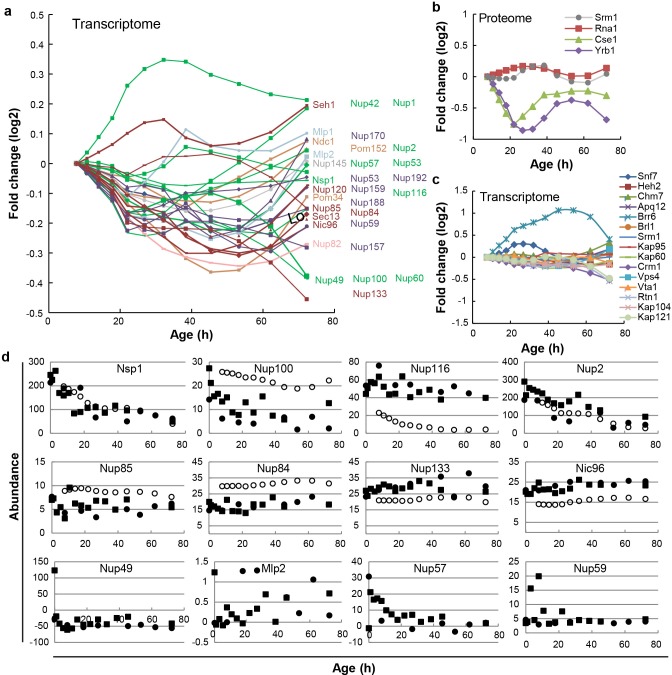
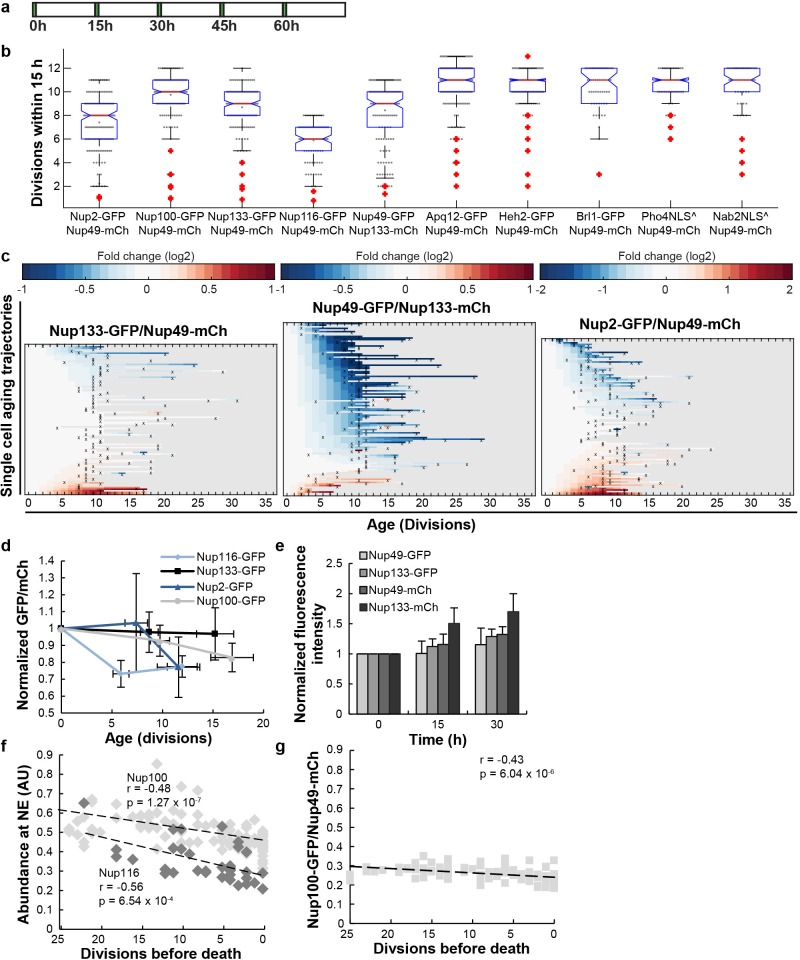
Figure 1. The cellular abundance of some NPC components changes in replicative aging.
(a) Cartoon representation of the NPC illustrates different structural regions of the NPC, all FG-Nups are shown in green independently of their localization, the membrane rings in light brown, the inner rings in purple, the outer rings in brown, the mRNA export complex in pink, and the nuclear basket structure in light blue. Adapted with permission from Springer Nature Customer Service Centre GmbH: Springer Nature, Nature, Integrative structure and functional anatomy of a nuclear pore complex, Kim et al. (2018). (b) Schematic presentation of replicative aging yeast cells. (c) Transcript and protein abundance of NPC components (color coded as in Figure 1a) as measured in whole cell extracts of yeast cells of increasing replicative age; after 68 hr of cultivation the average replicative age of the cells is 24. Cells were aged under controlled and constant conditions (Janssens et al., 2015). See also Figure 1—figure supplement 1a. (d) Young cells are trapped in the microfluidic device and bright field images are taken every 20 min to define the cells age and fluorescent images are taken once every 15 hr to detect the protein localization and abundance. Representative images of cells expressing indicated fluorescent protein fusions imaged at the start of the experiment and after 30 hr; their replicative age is indicated. Scale bar represents 5 µm. (e) Heat map representation of the changes in the levels of the indicated GFP- and mCh-tagged Nups at the NE in each yeast cell at increasing age. Each line represents a single cell’s life history showing the change in the ratio of the fluorescence from the GFP-tagged Nup over the fluorescence from the mCh-tagged Nup and normalized to their ratio at time zero. Measurement of the fluorescence ratios are marked with ‘x’; in between two measurements the data was linearly interpolated. The fold changes are color coded on a log 2 scale from −1 to + 1; blue colors indicate decreasing levels of the GFP-fusion relative to mCh. Number of cells in the heatmaps are Nup116-GFP/Nup49-mCh = 67, Nup133-GFP/Nup49-mCh = 94 and Nup100-GFP/Nup49-mCh = 126.
© 2018 Springer Nature
Figure 1A adapted with permission from Kim et al. (2018).