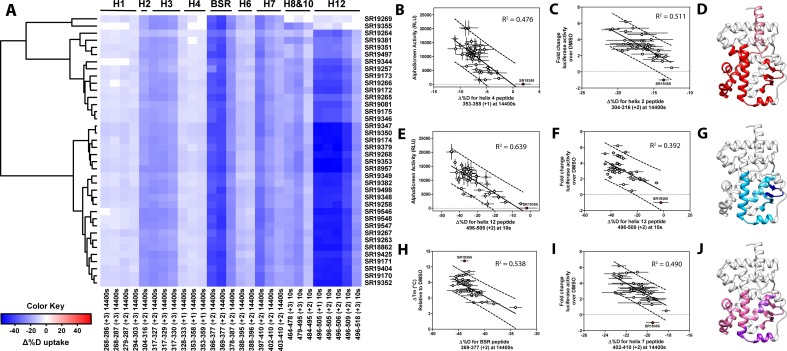
Figure 3. Differential HDX-MS and activity screening of 38 RORγ modulators reveals structural determinants for activation.
(A) Δ%D (difference from DMSO treated control) values for 38 RORγ modulators are plotted as a heat map according to the color key at the bottom left. Each row represents a compound and each column represents a peptide indicated by the labels at the right and bottom respectively. The locations of each peptide in the crystal structure are labeled along the top. A hierarchical clustering algorithm based on Ward’s method was performed to cluster compounds based on their exchange signatures and the corresponding dendrogram is shown on the left. (B-J) Activity correlation analysis results found that structural dynamics measurements correlate with functional activity. Regions of RORγ that correlate with coactivator peptide recruitment, thermal stability, and activation in cell-based assays are shown in panel D, G, and J, respectively. Lighter shades indicate that the slope of a linear regression was significantly (adjusted p value < 0.05) non-zero, while darker shades indicate an R (Takeda et al., 2012) was greater than 0.4. Representative coactivator peptide recruitment correlation plots for helix 12 and helix four are shown in B and E, respectively. The correlation between thermal stability and the BSR peptide are shown in panel (H). Representative correlation plots cell-based activity are shown for H2, H12 and H7 are shown in panels C, F, and I, respectively.