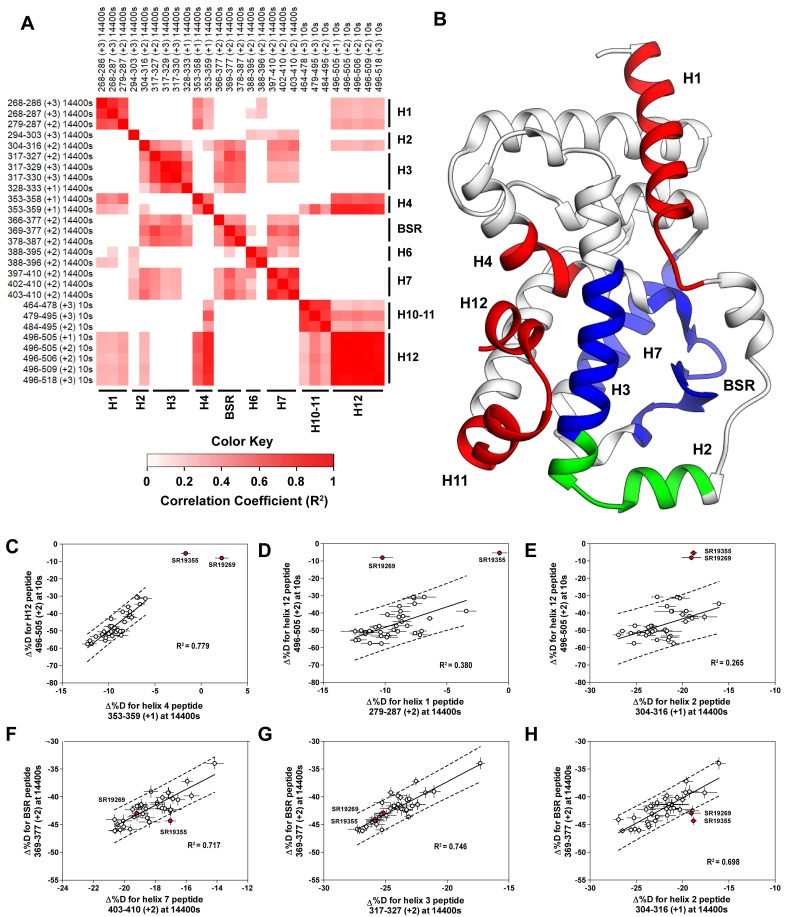
Figure 4. Covariation analysis of two timepoint HDX-MS screening reveals concerted ligand-dependent changes in protein dynamics.
(A) Correlogram showing correlation coefficients (R2) of Δ%D values between peptides. Peptides are labeled as start-end (charge) timepoint across the top and left sides of the panel while the location of the peptides are labeled along the bottom and right sides of the panel. Correlation coefficients that are significantly non-zero (adjusted P values < 0.01) slope are colored. (B) Perturbation values generally correlated in three groups. Group one correlated with H12 and consists of H1 and H4 and is colored red. Group two correlates with the BSR consists of H7 and H3 and is colored Blue. A peptide spanning residues 304–316 is shown in green. (C-H) Representative correlation plots showing distributions of compound perturbation values across peptides indicated by the x- and y-axis. Linear regression models are shown as solid black lines and the 95% confidence interval for prediction is shown as dashed lines. SR19355 and SR19269 (indicated in red) were often found as outliers for models involving helix 12 (panels C-E).