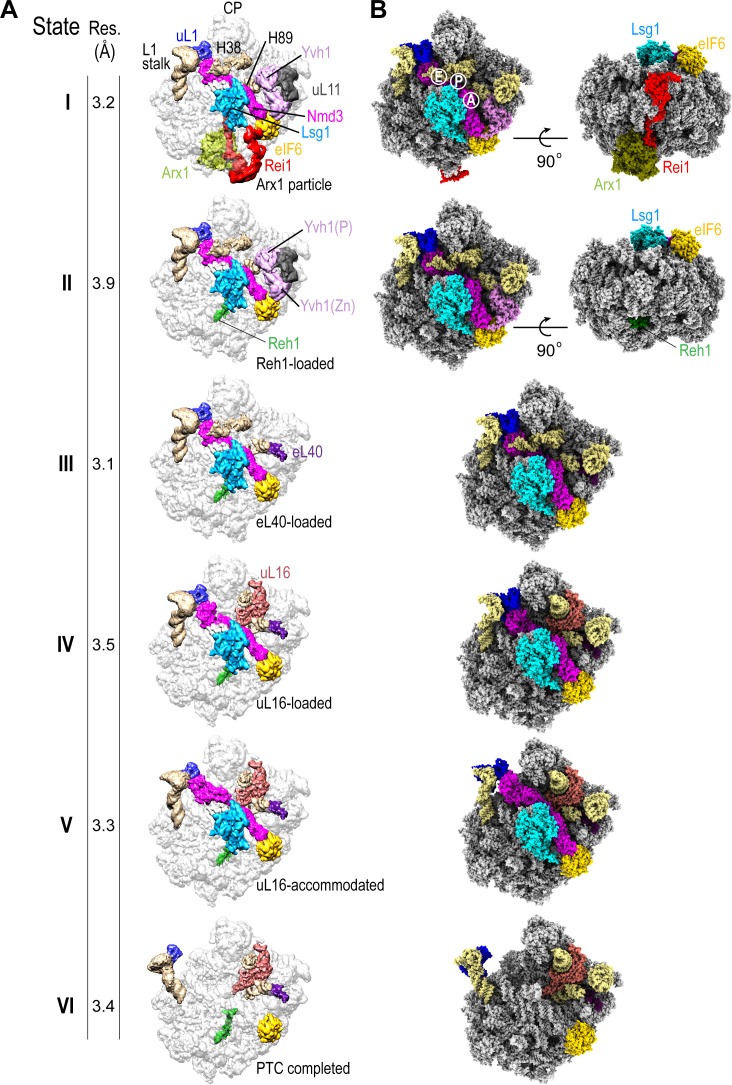
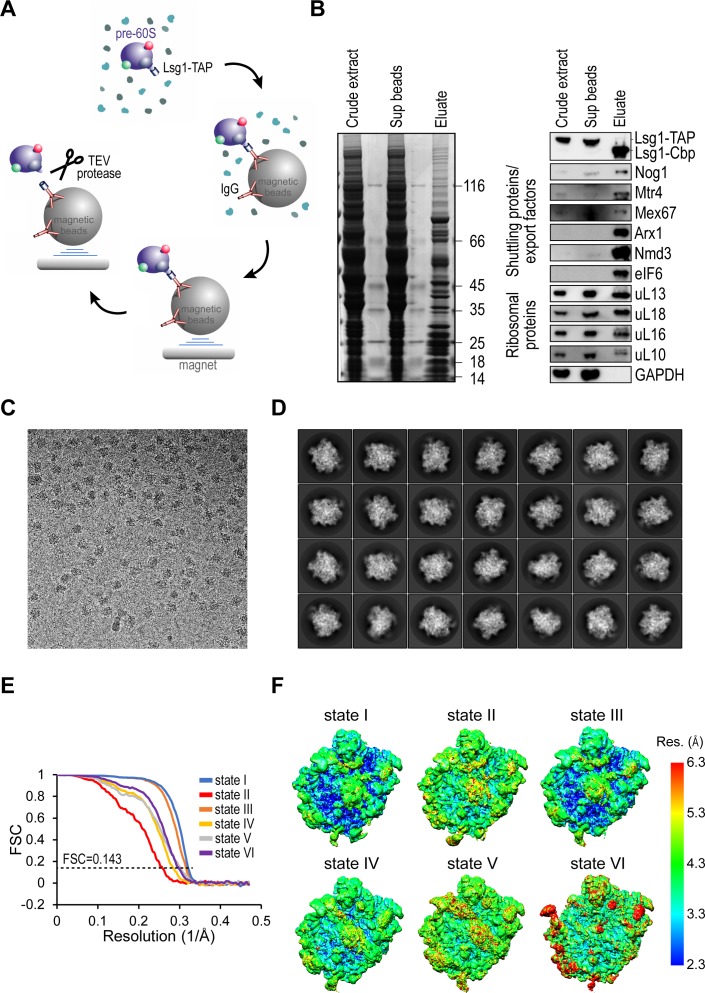
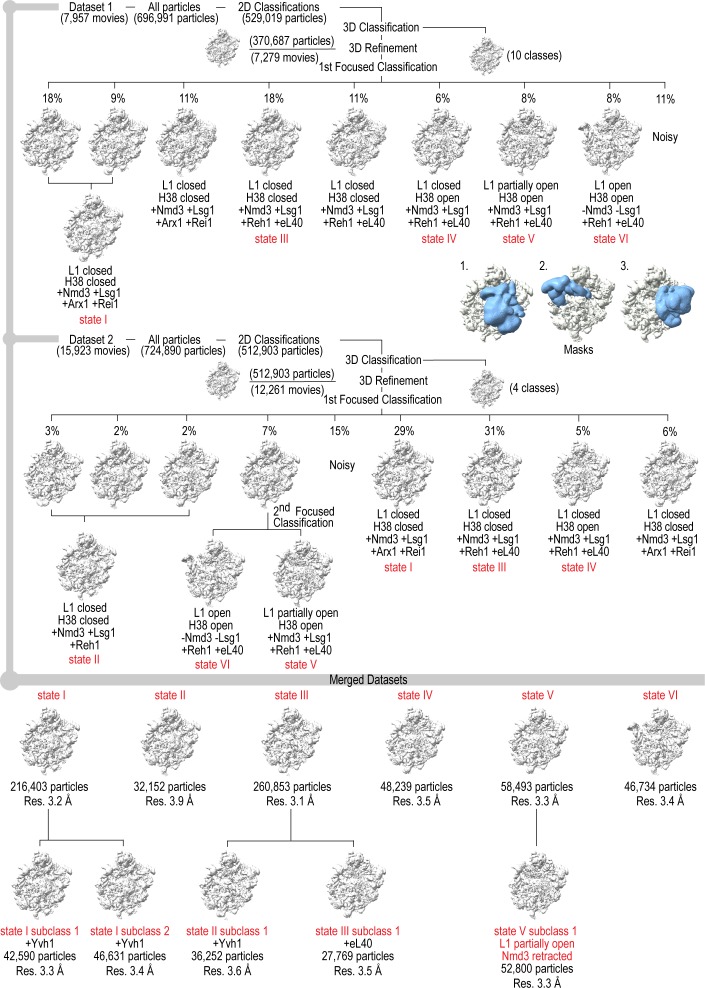
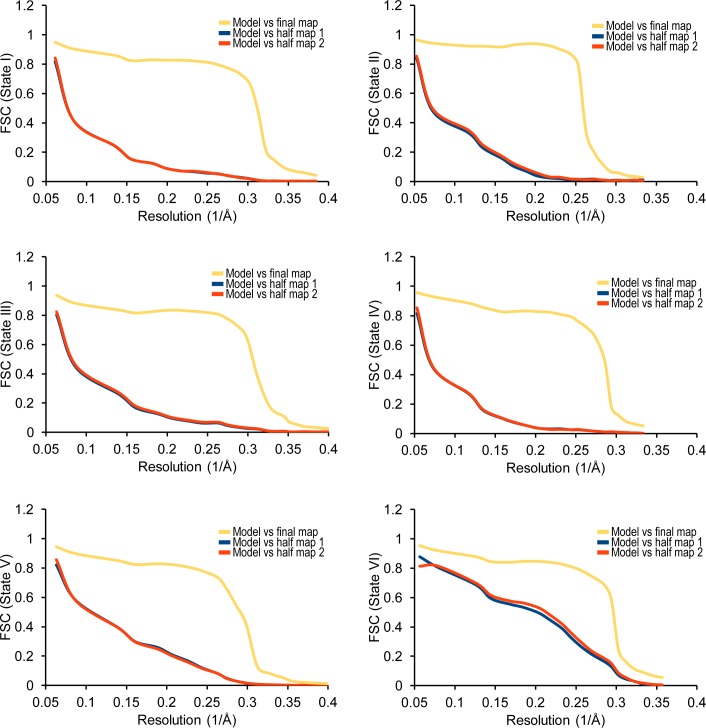
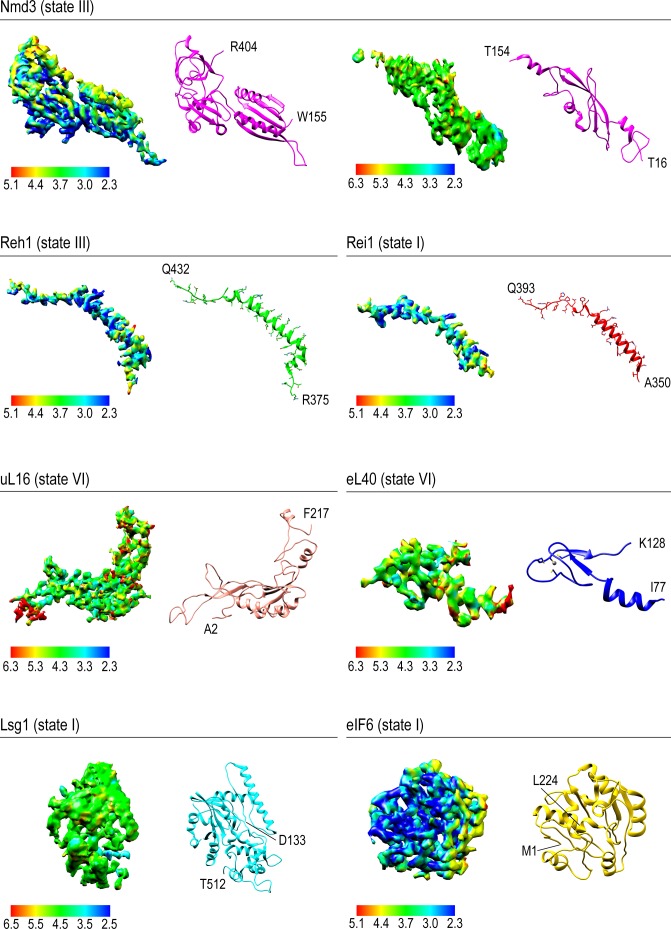
Figure 1. Sequential steps in late cytoplasmic 60S subunit maturation.
(A) Cryo-EM reconstructions of six cytoplasmic maturation states (I–VI). States, overall resolution, changes in protein composition and rRNA conformation are indicated. The phosphatase (P) and zinc finger (Zn) domains of Yvh1 are indicated in state II. (B) Atomic models of pre-60S states I-VI with rRNA and biogenesis factors highlighted.