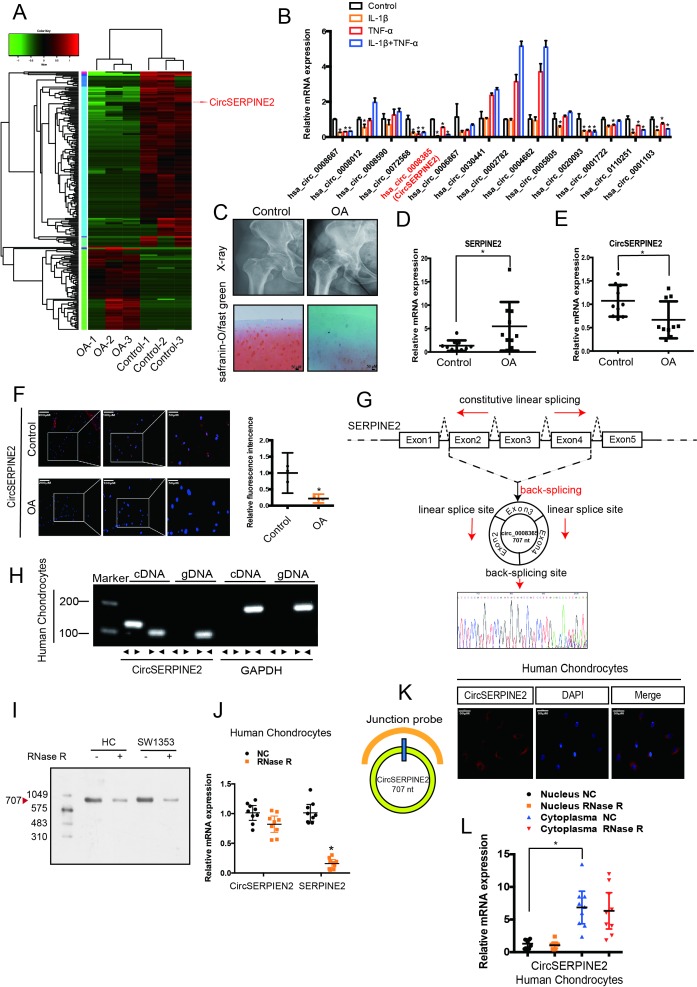
Figure 1.
CircSERPINE2 validation and expression in OA cartilage tissue and cells. (A) Heat map of all differentially expressed circRNAs between OA and control cartilage samples. (B) The expression of circRNAs in the chondrocytes stimulated with TNF-α (10 ng/mL) and IL-1β (10 ng/mL) for 24 hours. n=3 (three different experiments). *p<0.05. (C) X-ray (upper) and safranin-O/fast green (lower) staining of the cartilage from OA or control patients. Scale bar: 50 µm. (D) SERPINE2 expression was higher in human OA cartilage than in control cartilage. n = 10. *p<0.05. (E) CircSERPINE2 expression was lower in human OA cartilage than in control cartilage tissue. n = 10. *p<0.05. (F) CircSERPINE2 expression was lower in human OA cartilage than in control cartilage tissue. Representative images are shown (scale bar, 50–200 µm). n=3 (three different donors). *p<0.05, mean with 95% CI. (G) Schematic illustration showing SERPINE2 exons 2–4 circularisation to form CircSERPINE2 (black arrow). The presence of CircSERPINE2 was validated by RT-PCR, followed by Sanger sequencing. Red arrow represents head-to-tail CircSERPINE2 splicing sites. (H) The presence of CircSERPINE2 was validated in HCs by RT-PCR. Divergent primers amplified CircSERPINE2 from cDNA, but not from genomic DNA. GAPDH was used as a negative control. (I) Northern blot analysis for the detection of CircSERPINE2 expression in HC and SW1353 cells treated with or without RNase R digestion. The panels show the blots probed with CircSERPINE2, and the red triangle indicates CircSERPINE2 band size (707 nt). (J) The expression of CircSERPINE2 and SERPINE2 mRNA in HC cells treated with or without RNase R was detected by RT-qPCR. The relative levels of CircSERPINE2 and SERPINE2 mRNA were normalised to the values measured for mock treatment. n=9 (three different donors for three different experiments). *p<0.05. (K) RNA FISH revealed the predominant localisation of CircSERPINE2 in the cytoplasm. CircSERPINE2 probes were labelled with Cy-3. Nuclei were stained with DAPI. Scale bar, 50 µm. Upper panel: FISH with junction-specific probes indicative of the cellular localisation of CircSERPINE2. n = 3 (three different donors). Scale bars=50 µM. Lower panel: CircSERPINE2 expression was detected in different cell fractions. Nuclear and cytoplasmic RNAs were extracted, and junction primers were used for CircSERPINE2 detection. U6 was used as an internal control for nuclear RNA, whereas GAPDH served as the control for cytoplasmic RNA. n=9 (three different donors for three different experiments). *p<0.05. CircRNAs, circular RNAs; DAPI, 4′,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridisation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HCs, human chondrocytes; IL, interleukin; OA, osteoarthritis; RT-qPCR, real-time quantitative PCR; TNF, tumour necrosis factor.