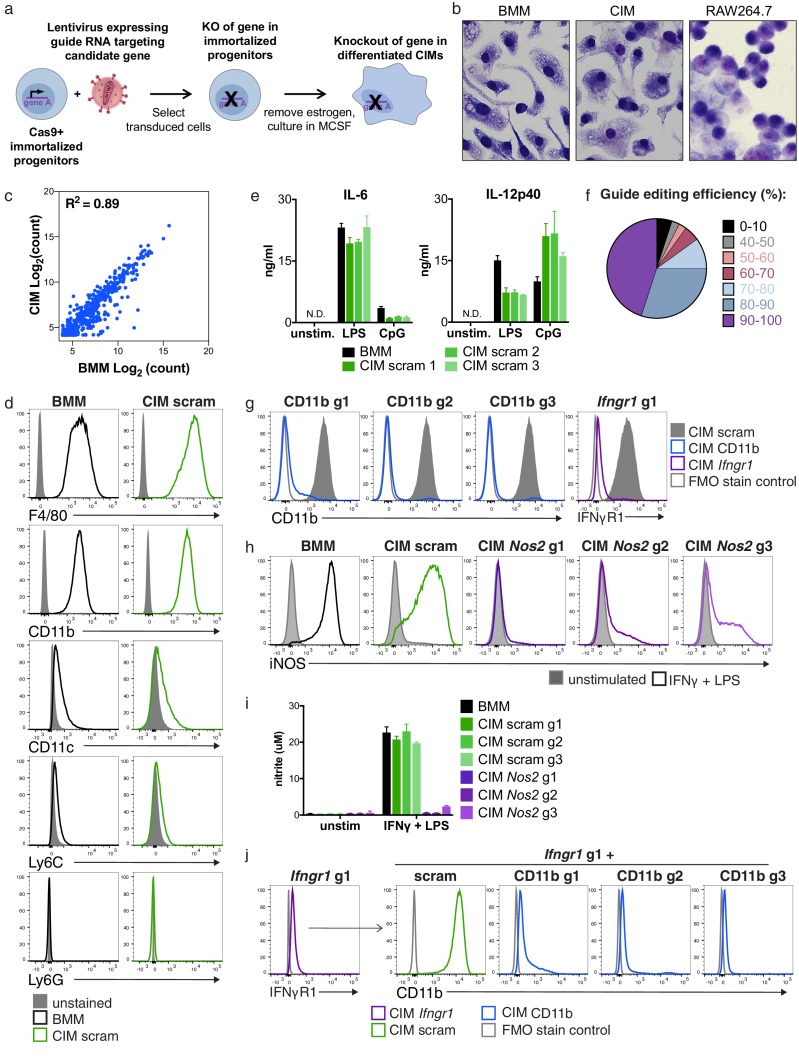
Figure 1. Cas9+ CIMs as a tractable system for genome-editing in macrophages.
(a) Graphic overview of gene editing in Cas9+ CIMs. (b) BMMs (left panel), CIMs (middle), or RAW 264.7 cells (right) were visualized with Diff-Quick stain. (c) mRNA levels in BMMs and CIMs was quantified using a Nanostring nCounter. Data are representative of two independent experiments and are presented as log transformed normalized transcript counts of the average of technical duplicates from one experiment. (d) BMMs or CIMs transduced with a scramble guide (CIM scram) were analyzed by flow cytometry for expression of the indicated myeloid cell markers. Data are representative of three independent experiments. (e) IL-6 and IL-12p40 production by BMMs or CIM scram stimulated with the TLR4 ligand LPS or the TLR9 ligand CpG was measured by ELISA. N.D. – none detected. Data are representative of three independent experiments each performed in triplicate, mean ± SD are shown. (f) Genomic DNA from CIMs transduced with 40 guides targeting 17 genes was analyzed for genomic editing by TIDE analysis. (g) CIMs transduced with a scramble guide or guides targeting CD11b or Ifngr1 were stained with the indicated antibodies. Fluorescence minus one (FMO) stained samples were used as controls. (h and I) BMMs or CIMs transduced with a scramble guide or guides targeting Nos2 were stimulated overnight with LPS + IFNγ or left unstimulated and then analyzed by flow cytometry for expression of iNOS (h); nitric oxide production in cell-free supernatants was analyzed by Griess assay (i). Data are representative of two independent experiments each performed in triplicate, mean ± SD are shown. (j) CIM progenitors previously transduced with a lentivirus containing puromycin resistance and a guide targeting Ifngr1 were subsequently transduced with lentivirus containing hygromycin resistance and scramble guide or guides targeting CD11b.