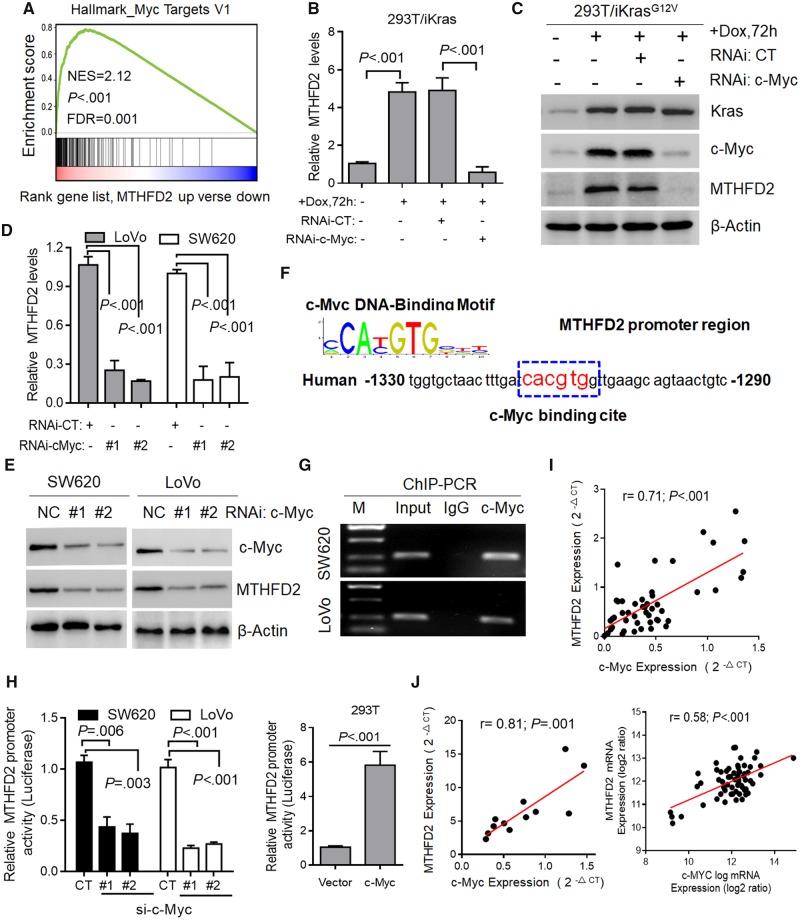
Figure 6.
Transcriptional regulation of methylene tetrahydrofolate dehydrogenase 2 (MTHFD2) in colorectal cancer (CRC). A) Gene set enrichment enrichment score and distribution of c-Myc-regulated genes along the rank of MTHFD2 up vs MTHFD2 down available from The Cancer Genome Atlas CRC database. B and C) Quantitative polymerase chain reaction (qPCR) and immunoblotting analysis of MTHFD2 expression in 293T/iKras cells after Kras induction and/or c-Myc depletion for 72 hours. D and E) qPCR and immunoblotting analysis of MTHFD2 expression in SW620 and LoVo cells transfected with c-Myc siRNAs (#1, #2). F) c-Myc DNA-binding sites are present in the human MTHFD2 promoter region. G) Chromatin immunoprecipitation-polymerase chain reaction (ChIP-PCR) in SW620 and LoVo cells. Representative of two independent experiments. H) Relative MTHFD2 luciferase promoter activity in indicated cells with c-Myc depletion or overexpression. I) Scatterplots of MTHFD2 vs c-Myc mRNA expression in CRC tissues (N = 48) analyzed by qPCR. J) Scatterplots of MTHFD2 vs c-Myc mRNA expression in cell lines (N = 62) available from CCLE database and CRC cell lines (N = 12). Pearson correlation coefficient (r) and P value are displayed. Data in B, D, and H are presented as the mean (SD) (n = 3), and statistical analyses were performed using Student unpaired t test. All statistical tests were two-sided. CT = control; DOX = doxycycline; NES = Normalized Enrichment Score; FDR = False Discovery Rate; NC = Negative Control; RNAi = RNA interference; IgG = Immunoglobulin G; si-c-Myc = Small interfering RNA-c-Myc.