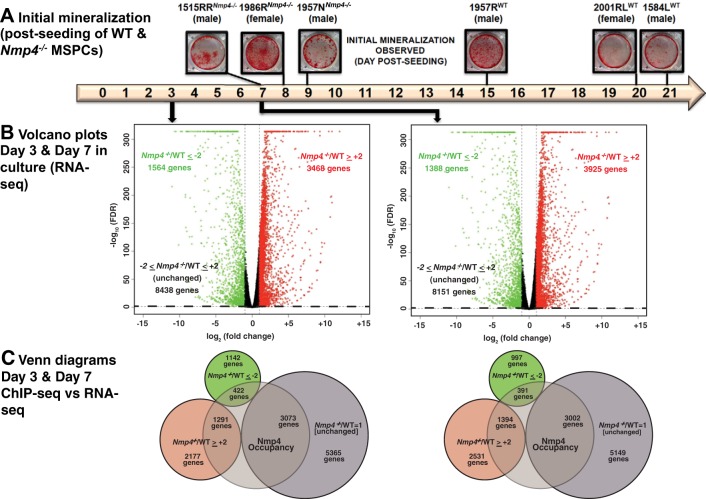
Fig. 1.
Loss of nuclear matrix protein 4 (Nmp4) accelerates and enhances mesenchymal stem/progenitor cell (MSPC) mineralization and has a broad impact on the transcriptome. A: 6 independently expanded MSPC preparations from individual wild-type (WT) and Nmp4−/− mice were established as described in materials and methods. Cultures were stained with alizarin red when mineralization was first observed. The cells derived from the Nmp4−/− mice consistently exhibited mineral days to weeks before this was observed in the WT cultures. The cell preparations 1957NKO and 1957RWT were derived from male littermates. The remaining lines were derived from male or female mice selected from random litters. B: volcano plots of RNA sequencing (RNA-seq) data from MSPCs maintained in nondifferentiating culture medium for 3 days and osteogenic differentiating culture medium for 7 days. The x-axis represents the logarithmic transformation to the base 2 of the mean fold change of mRNA expression in Nmp4−/− cells versus control cells and the y-axis represents the negative logarithm to the base 10 of the false discovery rate (FDR) value. Changes in gene expression were considered significant if the fold change of knockout (KO)/WT greater than or equal to +2 (red circles) and FDR ≤0.05 or KO/WT less than or equal to −2 (green circles) and FDR ≤0.05. The black circles represent genes that did not meet either criterion. The dotted line demarcates FDR = 0.05. C: Venn diagrams showing gene overlap between chromatin immunoprecipitation sequencing (ChIP-seq) and RNA-seq data. The former was derived from MC3T3-E1 cells (16). Genes that supported Nmp4 occupancy were required to exhibit peaks (height ≥10) within 5 to +2 kb from a transcription start site (TSS) and/or located within the range defined by the TSS and the transcription end site.