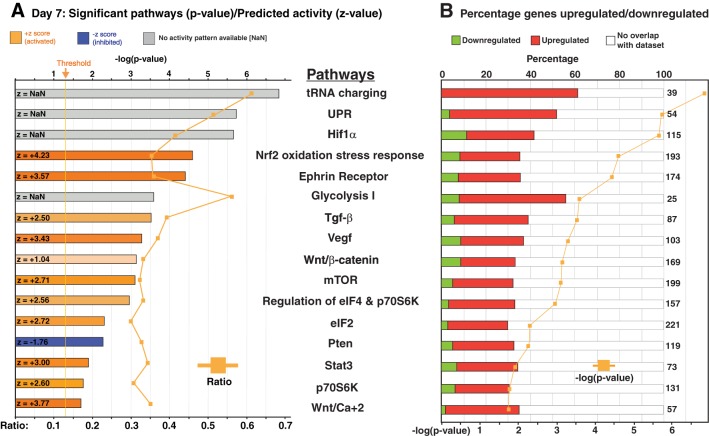
Fig. 2.
Ingenuity Pathway Analysis (IPA) of the RNA sequencing (RNA-seq) data identified over 200 pathways significantly altered in the nuclear matrix protein 4 knockout (Nmp4−/−) cells maintained in differentiation culture medium for 7 days (see Supplemental Table S4). Here we show select canonical pathways that are sensitive to Nmp4 status and relevant to the metabolic reprogramming, protein synthesis, and secretion of the bone cells. A: the bar graphs are color coded to reflect the z-score calculated by the IPA algorithm, which predicts the direction of change for the pathway upon loss of Nmp4. An absolute z-score of 2 or more is considered significant. The activation state of the pathway is predicted to be increased if the z-score is ≥2, and these bars are color coded with an orange hue. Conversely, bar graphs with a blue hue indicate a z-score less than or equal to −2 representing canonical pathways with a decreased activity. Those pathways represented with a gray bar (z = NaN) indicate that the z-score algorithm cannot predict whether the pathway activity is increased or decreased in the Nmp4−/− cells. The orange line gives the ratio of the number of genes listed in the Nmp4 data set over the total number of genes in the IPA-annotated pathway. B: the bar graphs are color coded to reflect the percentage of genes in a particular pathway whose expressions are significantly upregulated with the loss of Nmp4 (red) and those genes whose expressions are attenuated in the null cells (green). The total numbers of genes comprising the canonical pathways are also indicated. The orange line gives the –log10(P value), and significance was defined by P ≤ 0.05 [or 1.30 = −log10(P value)]. UPR, unfolded protein response; mTOR, mammalian target of rapamycin; Pten, phosphate and tensin homolog deleted on chromosome 10; eIF, eukaryotic initiation factor; Hif1α, hypoxia-inducible factor-1α; TGF-β, transforming growth factor-β.