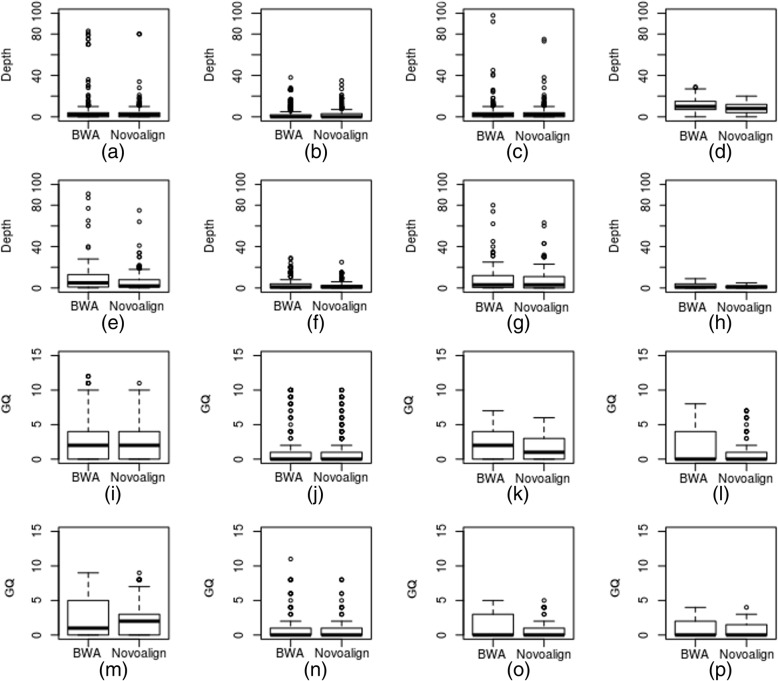
Fig. 7.
Analysis of depth and GQ of true SNVs missed (FN) by BWA and Novoalign alignments. Depth of the false negative SNVs on exome-1(a), − 2 (b), − 3(c) and − 4 (d) and InDels on exome-1(e), − 2(f), − 3(g) and − 4(h). Genotype quality of false negative SNVs on exome-1 to - 4 (i, j, k, and l) and InDels on exome-1 to − 4 (m, n, o and p) respectively