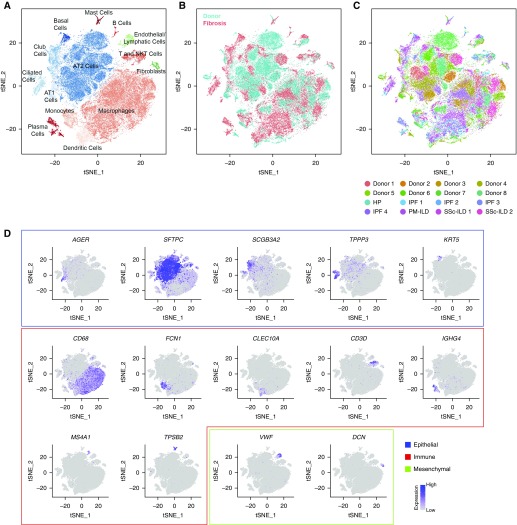
Figure 1.
Integrated single-cell RNA-Seq analysis of patients with pulmonary fibrosis identifies diverse lung cell populations. Single-cell RNA-Seq was performed on single-cell suspensions generated from eight lung biopsies from transplant donors and eight lung explants from transplant recipients with pulmonary fibrosis. All 16 samples were analyzed using canonical correlation analysis within the Seurat R package. Cells were clustered using a graph-based shared nearest neighbor clustering approach and visualized using a t-distributed Stochastic Neighbor Embedding (tSNE) plot. (A) Cellular populations identified. (B) Cells on the tSNE plot of all 16 samples were colored as originating either from a donor or from a patient with pulmonary fibrosis. (C) Each population included cells from donors and patients with pulmonary fibrosis. (D) Canonical cell markers were used to label clusters by cell identity as represented in the tSNE plot. Cell types were classified as epithelial, immune, or mesenchymal as indicated in the legend. AT1 = alveolar type I; HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; NK = natural killer; PM = polymyositis; SSc = systemic sclerosis.