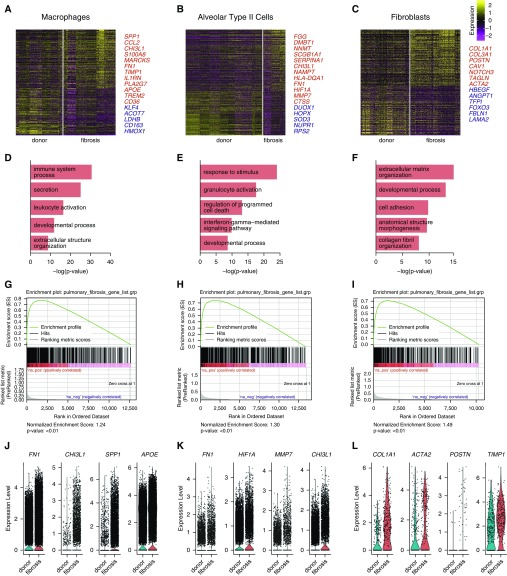
Figure 2.
Differential expression analysis of single-cell RNA-Seq data from normal and fibrotic lungs identifies genes characteristic of pulmonary fibrosis. (A–C) Differential expression analysis was performed comparing cells from normal and fibrotic lungs within macrophages, alveolar type II cells, and fibroblasts (Wilcoxon rank sum test as implemented within Seurat toolkit). Heatmaps are shown representing the upregulated and downregulated genes in macrophages, alveolar type II cells, and fibroblasts, highlighting genes involved in fibrosis. The full table of genes is found in Table E3. (D–F) Functional enrichment analysis with GO Biological Processes was performed using GOrilla with the significantly upregulated genes in cells from fibrotic compared with normal lungs. Representative significantly enriched GO processes are shown for macrophages, alveolar type II cells, and fibroblasts. (G–I) Gene Set Enrichment Analysis was performed using the Comparative Toxicogenomics Database Pulmonary Fibrosis Gene Set with genes ranked by log difference in average expression between fibrotic and normal lungs. Enrichment plots together with normalized enrichment scores and false discovery rate q values are shown for macrophages, alveolar type II cells, and fibroblasts. (J–L) Violin plots of expression for select genes significantly upregulated in patients with fibrotic compared with normal lungs.