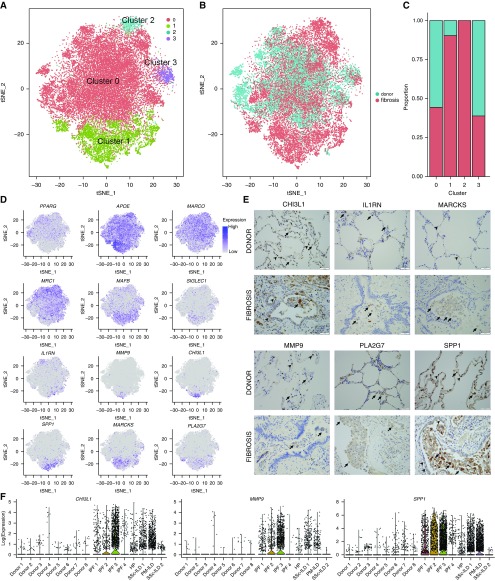
Figure 5.
Distinct populations of alveolar macrophages emerge during pulmonary fibrosis. (A) Cells identified as macrophages by individual annotation of single-cell RNA-Seq data from eight normal and eight fibrotic lungs were combined and then clustered, revealing four clusters. (B and C) Relative contributions of alveolar macrophages from normal and fibrotic lungs to each cluster as shown by t-distributed Stochastic Neighbor Embedding plot and by bar plots. (D) Feature plots demonstrating differential expression of selected alveolar macrophage maturation genes (PPARG, APOE, MARCO, MRC1, MAFB, and SIGLEC1), and genes associated with fibrosis (IL1RN, MMP9, CHI3L1, SPP1, MARCKS, and PLA2G7). (E) Immunohistochemistry on lung sections from the same patients confirms heterogeneity in fibrotic gene expression within alveolar macrophages. Arrows indicate positive staining in alveolar macrophages; arrowheads indicate positive staining in alveolar epithelium for CHI3L1 and SPP1, endothelium for MARCKS, and neutrophils for MMP9. Scale bars, 50 μm. (F) Violin plots representing heterogeneity in expression of CHI3L1, MMP9, and SPP1 in macrophages from donor and fibrotic lungs. HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; PM = polymyositis; SSc = systemic sclerosis; tSNE = t-distributed Stochastic Neighbor Embedding.