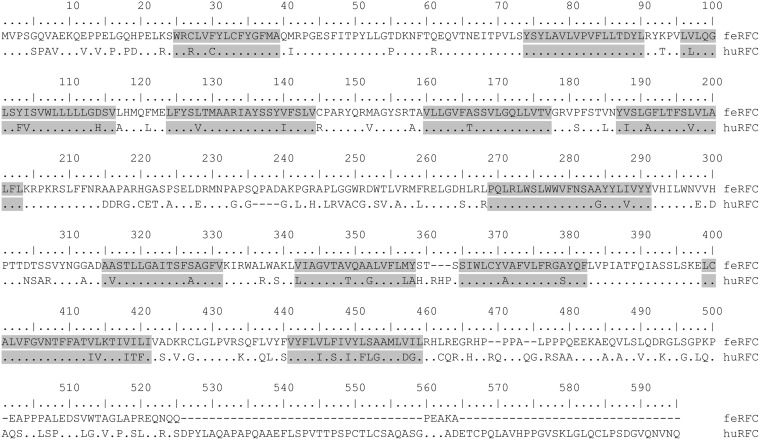
FIG 5.
Alignment of the amino acid sequences of feRFC and huRFC. The alignment in single letter amino acid code was conducted using MUSCLE software (66). Dots indicate conserved amino acid residues, and positions where there are differences between feline and human RFC sequences are shown as letters. Hyphens indicate gaps in the amino acid sequence. Transmembrane domains (gray boxes) based on huRFC were predicted using the constrained consensus topology prediction (74).