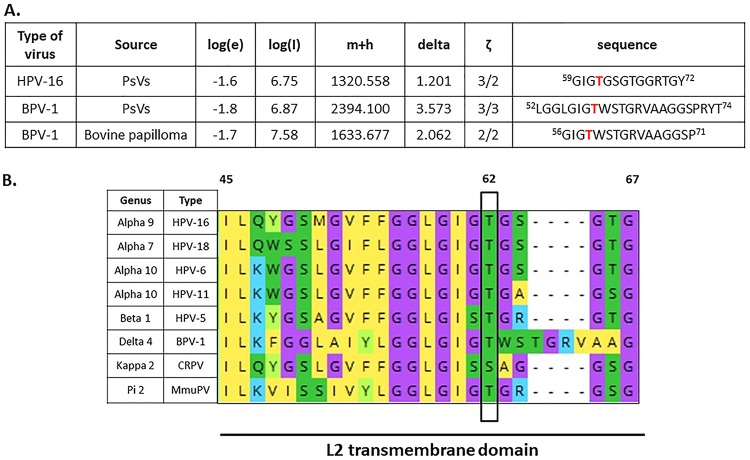
FIG 1.
Identification of the conserved phospho-acceptor site T62 in HPV-16 L2 capsid protein using mass spectrometry analysis. (A) The bicistronic plasmids expressing HPV-16 or BPV-1 L1 and L2 were transfected into HEK293TT cells, together with a luciferase reporter plasmid. After 48 h the cells were harvested, and pseudovirions were purified by cesium chloride gradient and analyzed by mass spectrometry. The spectra for L2 were analyzed for phospho-modifications and compared with those obtained from native BPV-1 virions isolated from a bovine papilloma. The threonine residue (T62 in HPV-16 L2 and T59 in BPV-1 L2) was phosphorylated in both PsVs and native virus. The table shows results from the mass spectrometric analysis: log(e), the base 10 log of the expectation that the assignment is stochastic; log(l), the base 10 log of the sum of the intensities of the fragment ion spectra; m+h, the calculated mass of the protonated parent ion for this sequence assignment; delta, the difference between the measured and calculated protonated parent ion masses; ζ, the ratio of the measured charge of the parent ion to the number of basic sites in the assigned peptide sequence. The sequence of the assigned peptide is also shown. (B) Alignment of L2 transmembrane (TM) domains of selected PVs. Alignment was generated with Mega X software. Conserved threonine/serine phospho-acceptor (T62/S62) sites in the L2 sequences are highlighted in green and boxed. Note that residue T62/S62 is highly conserved across a wide evolutionary spectrum of PV types, ranging from high- and low-risk HPV types to BPV-1, MmuPV, and CRPV-1.