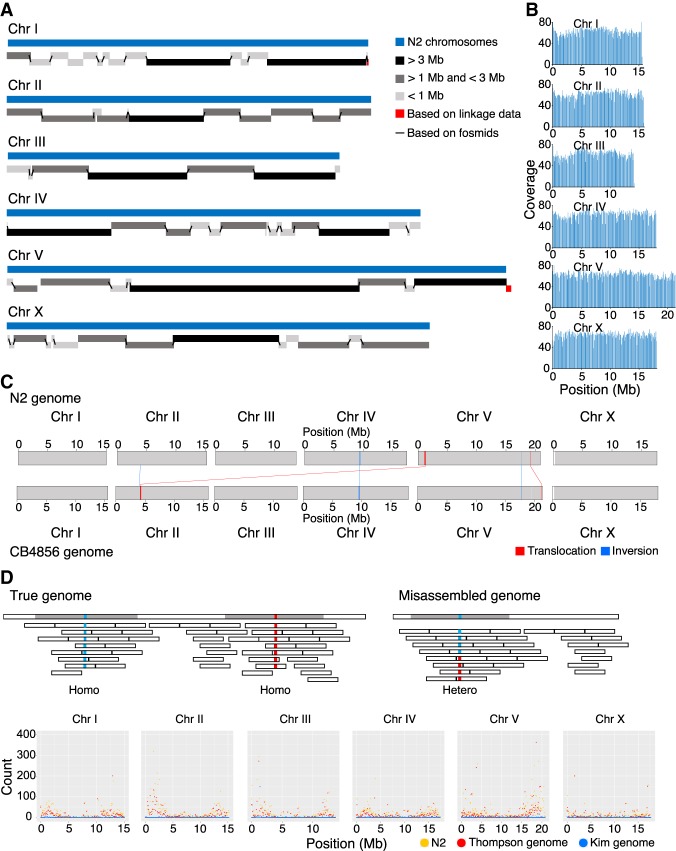
Figure 1.
CB4856 genome assembly and comparison with the N2 genome at a chromosome level. (A) Schematic representation of CB4856 contig lengths mapped to N2 WBcel235 chromosomes. (B) PacBio raw read coverage, mapped on CB4856 chromosomes (100-kb binned). Reads were distributed at average 60× coverage. (C) Schematic of large chromosomal rearrangement between N2 and CB4856 genomes identified using progressiveMauve. The blue box and line indicate inversion; the red box and line, translocation; and the white box indicates the unaligned block. Chr VR has several small rearrangements and unaligned blocks. Chr II: 3,896,126–3,900,949 in N2 was inverted in CB4856 (Chr II: 4,045,653–4,040,823), Chr V: 17,616,880–17,623,484 in N2 was inverted in CB4856 (Chr V: 17,734,209–17,728,873), and Chr V: 19,258,912–19,289,935 in N2 was located at Chr V: 21,193,104–21,237,336 in CB4856. (D) Schematic representation of CB4856 HiSeq reads mapped on the CB4856 genome (blue) or the N2 genome (yellow). Each dot shows the heterozygous base count (100-kb interval) from Chr I to Chr X.