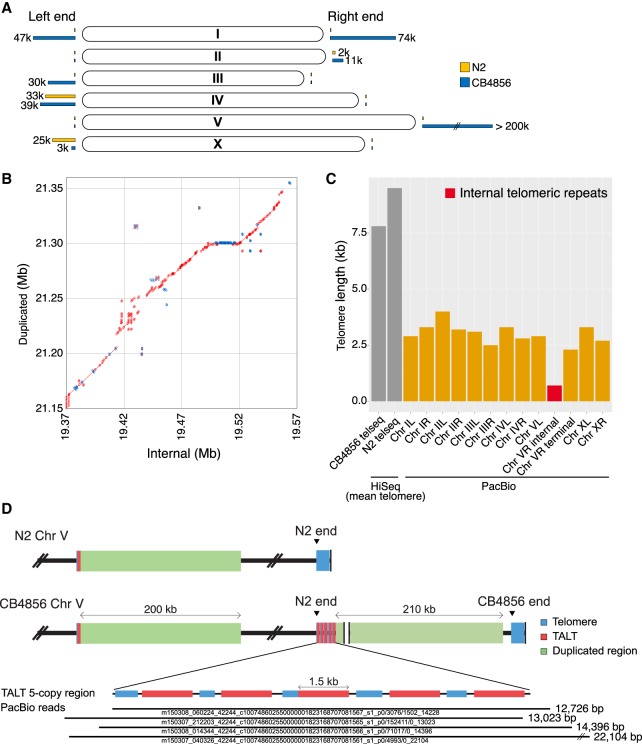
Figure 3.
New subtelomere formation in CB4856 Chr VR using an alternative lengthening of telomeres (ALT) mechanism. (A) Schematic representation of subtelomere differences between the N2 and CB4856 chromosomes. Yellow bars and blue bars at the end of chromosomes indicate the ratio of unaligned bases of subtelomeres in N2 and CB4856 genome, respectively. (B) Dot plot representing alignment between internal segment (V: 19,377,978–19,606,221) and duplicated segment (V: 21,171,521–21,389,866) of CB4856 Chr VR; 63% of the two regions are aligned, and 91% of the aligned bases are identical. Red: forward strand matches; blue: reverse strand matches. (C) Telomere length of all chromosomes deduced from the long-read CB4856 genome. ‘HiSeq’ data are mean telomere lengths normalized by the telseq software (Ding et al. 2014). The red bar represents the end of N2 (Chr VR internal) in Chr VR of CB4856. Only small portions of the N2 telomere remain in CB4856, followed by a new subtelomere. ‘Chr V terminal’ is from the real end of Chr VR. (D) Schematic representation of Chr V subtelomere in CB4856. Five copies of template for ALT (TALT) (red) are connected to the duplicated segment from the internal segment close to the internal TALT (V: 19,366,148–19,367,611). The bottom shows PacBio raw reads on the tandemly repeated TALT region. Four raw reads almost fully cover this region.