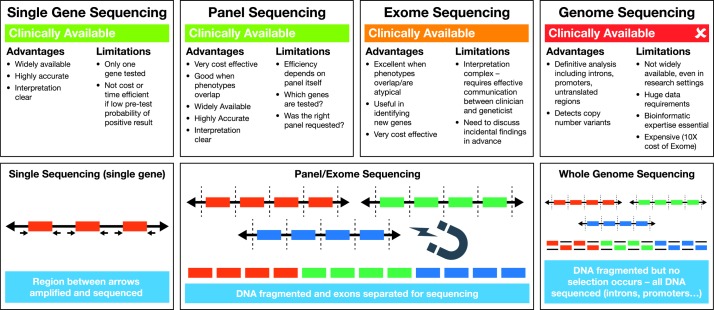
Figure 3.
A schematic diagram to illustrate currently available sequencing technologies. Single-gene (Sanger) sequencing is widely available and is performed by amplifying coding regions of the gene of interest by PCR, before sequencing these regions. Introns and promoter regions are not routinely sequenced. Next-generation sequencing-based panel or exome sequencing is becoming more widely available clinically. In this technique, DNA is fragmented, and the exons of multiple genes of interest are selected out for sequencing. Although many genes are sequenced, again only the coding regions are selected. In whole genome sequencing, which is not routinely available to clinicians yet, the DNA is also fragmented, but all the resulting DNA is sequencing, without a selection step. This results in a large amount of data, as the coding regions, intergenic regions, introns and promoters of all human genes are sequenced. Advanced bioinformatics and computing technology are a required part of whole genome sequencing.