Abstract
Genome-wide association studies (GWASs) have identified >100 susceptibility loci for schizophrenia (SCZ) and demonstrated that SCZ is a polygenic disorder determined by numerous genetic variants but with small effect size. We conducted a GWAS in the Japanese (JPN) population (a) to detect novel SCZ-susceptibility genes and (b) to examine the shared genetic risk of SCZ across (East Asian [EAS] and European [EUR]) populations and/or that of trans-diseases (SCZ, bipolar disorder [BD], and major depressive disorder [MDD]) within EAS and between EAS and EUR (trans-diseases/populations). Among the discovery GWAS subjects (JPN-SCZ GWAS: 1940 SCZ cases and 7408 controls) and replication dataset (4071 SCZ cases and 54479 controls), both comprising JPN populations, 3 novel susceptibility loci for SCZ were identified: SPHKAP (Pbest = 4.1 × 10−10), SLC38A3 (Pbest = 5.7 × 10−10), and CABP1-ACADS (Pbest = 9.8 × 10−9). Subsequent meta-analysis between our samples and those of the Psychiatric GWAS Consortium (PGC; EUR samples) and another study detected 12 additional susceptibility loci. Polygenic risk score (PRS) prediction revealed a shared genetic risk of SCZ across populations (Pbest = 4.0 × 10−11) and between SCZ and BD in the JPN population (P ~ 10−40); however, a lower variance-explained was noted between JPN-SCZ GWAS and PGC-BD or MDD within/across populations. Genetic correlation analysis supported the PRS results; the genetic correlation between JPN-SCZ and PGC-SCZ was ρ = 0.58, whereas a similar/lower correlation was observed between the trans-diseases (JPN-SCZ vs JPN-BD/EAS-MDD, rg = 0.56/0.29) or trans-diseases/populations (JPN-SCZ vs PGC-BD/MDD, ρ = 0.38/0.12). In conclusion, (a) Fifteen novel loci are possible susceptibility genes for SCZ and (b) SCZ “risk” effect is shared with other psychiatric disorders even across populations.
Keywords: polygenic score, SLC38A, SPHKAP, CABP1, ACADS, GWAS
Introduction
A recent meta-analysis of genome-wide association studies (GWASs), conducted by the Schizophrenia Working Group of the Psychiatric Genomics Consortium (PGC-SCZ) has highlighted more than 100 susceptibility loci for schizophrenia (SCZ).1 However, due to the extremely small effect size of the susceptibility variants, many SCZ loci remain undetected2; this finding is also applicable to other psychiatric disorders, including bipolar disorder (BD)3,4 and major depressive disorder (MDD).5–8
Numerous studies examining polygenic architecture based on a polygenic risk score (PRS) analysis have supported the “small effect size” of the susceptibility variants for psychiatric disorders. The PRS analysis revealed that the occurrence of SCZ and/or other psychiatric disorders, including BD and MDD, is explained by the polygenic model, where numerous genetic variants with small effect sizes accumulatively influence the liability of the psychiatric disorders. In detail, the PGC-SCZ reported that the explained-variance in SCZ liability was estimated to be approximately 15%, based on the “training” PGC-SCZ dataset on the “target” other SCZ GWAS results.1 In that study, the “risk” single-nucleotide polymorphisms (SNPs) identified in SCZ populations with European (EUR) ancestry were also shared with SCZ detected in other populations such as African Americans (AA) and East Asians (EAS). Furthermore, PRS analysis suggests that common genetic features exist beyond diagnostic boundaries in psychiatric disorders,9 but, as expected, the explained-variance in “trans-disease” analysis findings substantially decreased compared with “identical-disease” analysis findings. This evidence is supported by the use of a different method, genetic correlation analysis such as the linkage disequilibrium (LD) score regression analysis,10 indicating that the genetic effect is substantially correlated among SCZ, BD, MDD, and other psychiatric disorders.10,11
However, the data of “trans-disease/population analyses” for determining the PRS and/or genetic correlations between disorders across populations remain limited, mainly because of the small number of GWASs of non-EUR samples, and such studies tended to target a small sample size (1836 SCZ/3383 controls in PGC1) compared to EUR data (33640 SCZ/43456 controls in PGC1). This “trans-population” analysis can provide insightful speculations such as the presence of a shared genetic risk of SCZ and/or between psychiatric disorders across populations, even though different properties of LD and genetic variants with low minor frequencies exist in different populations.12
In this study, to identify novel SCZ susceptibility genes, we performed the GWAS of SCZ in a Japanese (JPN) population and a meta-analysis of data acquired from the PGC-SCZ and Chinese SCZ GWASs. Furthermore, we examined the shared genetic risk of SCZ and/or of psychiatric disorders across populations.
Methods
Subjects
Discovery GWAS Subjects (JPN-SCZ GWAS).
The discovery GWAS (Illumina Human OmniExpressExome v1.0, Illumina) initially included 2131 SCZ subjects (diagnosed by DSM-IV-TR, age mean ± standard error (SD): 48.0 ± 14.4, male/female ratio = 0.50/0.50), with 1940 SCZ subjects remaining after genotype quality control (QC). The comparison subjects were nonpsychiatric controls recruited for the BioBank Japan. These JPN-SCZ GWAS controls (7408 subjects genotyped by Illumina Human OmniExpress v1, age: 58.7 ± 13.4, male/female ratio = 0.58/0.42: supplementary table S1) were entirely identical to those used in our previous Phase I GWAS4 of BD (JPN-BD GWAS comprised 2 datasets: Phase I [cases/controls] and Phase II [cases/controls]).
It is of note that the case samples (243 SCZ cases in the JPN-SCZ GWAS) comprised half of the SCZ subjects used in our previous paper13; therefore, we could not conduct a simple meta-analysis of current and previous13 “JPN-GWAS” data. For the meta-analysis with PGC data, the previous Japanese GWAS was involved in the PGC (243 SCZ were overlapped), therefore we used only PGC-SCZ EUR data for the meta-analysis of current and PGC data, as mentioned below.
Replication Subjects for the “Top Hits” SNPs (JPN-SCZ REP).
The replication case samples consisted of 4189 SCZ subjects (diagnosed by DSM-IV-TR, age: 45.4 ± 14.7, male/female ratio = 0.53/0.47, supplementary text), with 4071 SCZ remaining after genotype QC, and the comparison group included 54479 subjects (age: 60.9 ± 14.8, male/female ratio = 0.57/0.43) who had also been genotyped (Illumina Human OmniExpressExome v1.2) as case subjects for 14 nonpsychiatric disorders (supplementary table S1). These comparison subjects were also identical to those used in our Phase II GWAS of BD.4
Genotyping, QC, and Imputation
Detailed QC, including population stratification (supplementary figure S1), is presented in the supplementary text. We then performed genotype imputation using a subset of the 1000 Genomes Project Phase I, which comprised the Japanese in Tokyo (JPT), Han Chinese in Beijing (CHB), and Southern Han Chinese (CHS) populations14 as a reference (supplementary text). After imputation, we included only SNPs with an imputation quality score of R2 ≥ 0.3 and a minor allele frequency of >1%.
For replication analysis of the “top hit” SNPs with the arbitrary threshold (P-value <1 × 10−4), the replication SCZ subjects (JPN-SCZ REP) were genotyped through custom typing using HiSeq2500 (Illumina: details can be seen in the supplementary text). First, we assessed overlapping or related (within 2 degrees) samples using genotype data by an identity-by-state analysis and removed 118 cases. For the SNP-wise QC, 86 SNPs did not pass our QC (exclusion criteria: call rate < 0.99 or P-value in Hardy–Weinberg Equilibrium < 1 × 10−6 for genotyped SNPs and imputation R2 ≤ 0.90 in the controls for imputed SNPs). Accordingly, 271 SNPs were candidates for the replication analysis (genotypes in 4071 SCZ vs imputations in 54479 controls).
Statistical Analysis
GWA Analysis and Meta-analysis.
For the SNP-based association analysis, we performed logistic regression using the mach2dat program, with the allele dosage as an explanatory variable and the first 2 eigenvectors (calculated based on Japanese Mainland [Hondo cluster]15: supplementary text) as independent variables.16,17 We calculated the combined P-values of the “discovery” JPN-SCZ GWAS, replication results for the “top hit” SNPs (JPN-SCZ REP), PGC-SCZ data, and recent GWAS of the Chinese population,18 using a fixed-effect model or Han and Eskin’s Random Effects model (RE2: if heterogeneity P < .05), which is suitable for the analysis of heterogeneous samples.19 Briefly, we assessed the following datasets: (a) JPN-SCZ GWAS (1940 SCZ/7408 controls); (b) JPN-SCZ GWAS and JPN-SCZ REP (4071 SCZ/54470 controls) (total 6011 SCZ/61878 controls); (c) JPN-SCZ GWAS and PGC-SCZ in EUR (PGC-SCZ EUR: 33640 SCZ/43456 controls) (total 35580 SCZ/50864 controls); (d) JPN-SCZ GWAS and Chinese SCZ GWAS (CHN-SCZ GWAS: 7699 SCZ/18327 controls)/Replication (CHN-SCZ REP: 4384 SCZ/5770 controls) (total 14023 SCZ/31505 controls); and (e) JPN-SCZ GWAS, CHN-SCZ GWAS/CHN-SCZ REP, and PGC-SCZ EUR (total 47663 SCZ/74961 controls). Regarding the combined analysis of CHN-SCZ GWAS/CHN-SCZ REP, only the whole-genome results of the final meta-analysis between the CHN-SCZ GWAS and the PGC-SCZ EUR, not the results of the “Chinese only” association analysis except their top hits, were provided. Therefore, we highlighted the analysis of the top hits for “Chinese only” results listed in the paper18 (corresponding to analysis d) as well as genome-wide SNPs of the meta-analysis among our JPN-SCZ GWAS, CHN-SCZ GWAS, and PGC-SCZ EUR (corresponding to analysis e). The details of the overall workflow for the combinations of the association analyses can be seen in supplementary figure S2.
To assess novel loci for the Japanese samples, which were not reported as the “108” PGC-SCZ regions,1 a clumping procedure in accord with the PGC-SCZ paper1 was carried out, although some of the Japanese samples (N = 243 SCZ) were involved in PGC-SCZ data, mentioned above: using PLINK1.9 (https://www.cog-genomics.org/plink/1.9/. Accessed September 25, 2018), with R2 at 0.1 within 500-kb chunks, based on the Phase 1 (ASN) dataset of the 1000 Genomes Project.14 Regional association plots were generated using LocusZoom20 or ggplot2 software in R. The significance level was set at 5 × 10−8 (2-sided).
Additional Bioinformatics Analysis.
We also conducted (a) HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php. Accessed September 25, 2018) analysis to evaluate the functional annotation of significant SNPs; (b) MAGMA21 analysis (v1.06) for pathway analysis. Details provided in the supplementary text.
PRS Analysis.
For the PRS analysis,1,2,22 the statistical analysis software PRSice v1.2 was used.23 The P threshold (PT) for selecting the “risk” SNPs was set sequentially at 0.1, 0.2, 0.3, 0.4, and 0.5; SNPs were selected if their P-values were between 0 and the chosen value of PT. Given that the inclusion of SNPs in the major histocompatibility complex (MHC) region could inflate the score due to high LD, we removed SNPs in this region by setting the “remove.mhc” flag in the software. The eligible SNPs for PRS were then selected based on LD-clumping (performed using the default setting of the software). The explained variance for the PRS was estimated using Nagelkerke’s R2 in a logistic regression model.
All comparison subjects in this study (JPN-SCZ GWAS) were shared with those in the JPN-BD GWAS (this comprised 2 datasets: Phase I [cases/controls] and Phase II [cases/controls]). Specifically, Phase I controls were identical to the controls in JPN-SCZ GWAS; therefore, to calculate the PRS between SCZ and BD within the JPN population only, we compared the data between the JPN-SCZ GWAS and the Phase II case-control GWAS (1419 BD cases vs 54479 controls) of JPN-BD, which was independent of Phase I controls.4
As discovery datasets, publicly available GWAS data for SCZ (PGC-SCZ EUR),1 BD (PGC-BD [EUR]),3 MDD (PGC-MDD [EUR]8 and CONVERGE [EAS]7) and our own data for BD (JPN-BD [EAS]4) were used (SNPs with R2 or INFO ≥ 0.6, supplementary table S2).
The significance level for the PRS analysis was set at 0.001, a conservative threshold described by Euesden et al.23
LDSC Analysis.
We calculated SNP heritability24 and assessed the “polygenicity”25 by LD score regression. In this analysis, we used LDSC software and pre-computed LD scores for EAS and EUR populations, based on the 1000 Genomes Project data listed on the website (https://data.broadinstitute.org/alkesgroup/LDSCORE/eas_ldscores.tar.bz2. Accessed September 25, 2018). To calculate the genetic correlation (rg) among subjects within the same population (“EAS vs EAS” or “EUR vs EUR”), we also used LDSC software and filtered the SNPs to HapMap3 SNPs (“--merge-alleles” flags).
To obtain the genetic correlation among subjects from different populations (ie, comparison between EAS and EUR), we used Popcorn software (v.0.9.6), which is based on a concept similar to the LD score regression,12 because LD score regression can assess genetic correlations only when samples from the same population are targeted. We examined (a) the trans-population genetic effect correlation (the correlation coefficient for the per-allele SNP effect sizes, ρge) and (b) the genetic impact correlation (the correlation coefficient for the population-specific allele variance normalized SNP effect sizes, ρgi).12 In this analysis, we used HapMap3 datasets, representing the EUR population by the CEU dataset and the Asian population by CHB+JPT datasets. Genetic correlations were then determined according to the GWAS SNPs (with R2 or INFO≥0.6) in the EAS and EUR using the default setting, which removed SNPs below the minor allele frequency cutoff of 5% and those with A/T or G/C alleles.
To assess the trans-disease correlation (within the same population or across populations), we also used the aforementioned publicly available GWAS data (supplementary table S2).
Results
SCZ GWAS of the JPN Dataset (JPN-SCZ GWAS and JPN-SCZ REP)
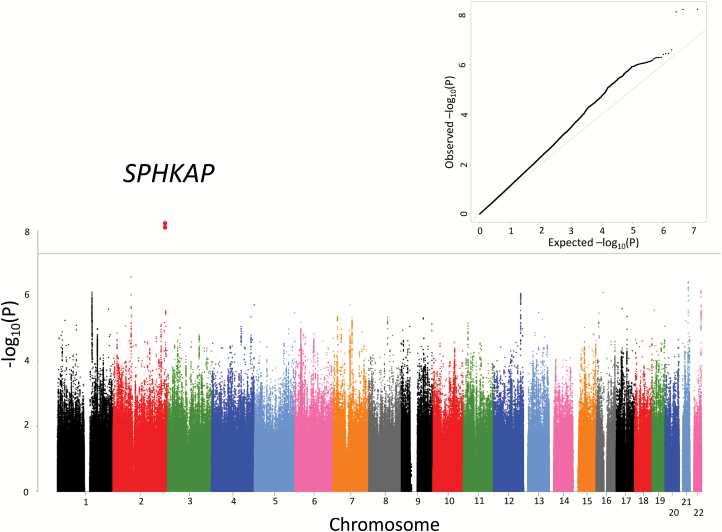
A total of 6627481 imputed SNPs in 1940 SCZ and 7408 nonpsychiatric subjects passed our stringent QC. The Manhattan and quantile–quantile plots based on these results are shown in figure 1. We observed that the genomic inflation factor (λGC) was at 1.03 and the intercept from the LDSC analysis was at 1.084, indicating that most of the inflation was due to polygenicity rather than bias.25
Fig. 1.
Manhattan and quantile–quantile plots of the JPN-SCZ GWAS. Horizontal line in the Manhattan plot indicates the threshold for genome-wide significance (P < 5 × 10−8). JPN, Japanese population; SCZ, schizophrenia; GWAS, Genome-Wide Association Study.
One region reached genome-wide significance (5 × 10−8; figure 1) at a SNP (rs10204933) in the SPHK1 interactor, AKAP domain-containing gene (SPHKAP; P = 6.3 × 10−9; OR = 1.63; table 1). In addition, we detected a number of “marginal” associations (P < 1 × 10−6) with SCZ (supplementary table S3). Interestingly, SNPs in the MHC region, for which a previous GWAS in a EUR population revealed a definitive association,1 showed a nonsignificant trend for the association (SNP with the best P-value: rs2393910, chr6:27057077, P = 6.3 × 10−6; OR = 0.76; however, in the PGC-SCZ EUR results, this SNP was associated with SCZ in the EUR samples but with the opposite direction of the effect [P = 1.8 × 10−5; OR = 1.07]).
Table 1.
Meta-analysis Between JPN-SCZ GWAS and JPN-SCZ Replication Datasets
| SNP | CHR | BP | Closest Gene | A1/A2 | Meta-analysis (JPN-SCZ GWAS+JPN-SCZ REP) | JPN-SCZ GWAS (1940 case/7408 control) | JPN-SCZ REP (4071 Case/54479 Control) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CIs | P-value | Case Freq. (A1) | Control Freq. (A1) | OR | P-value | Case Freq. (A1) | Control Freq. (A1) | OR | P-value | |||||
| rs10204933 | 2 | 229071614 | SPHKAP | G/A | 1.30 | 1.21–1.39 | 4.11.E-10 | 0.058 | 0.036 | 1.63 | 6.3.E-09 | 0.052 | 0.044 | 1.19 | 6.7.E-04 |
| rs12928842 | 16 | 84077987 | SLC38A8 | C/T | 1.14 | 1.10–1.18 | 5.68.E-10 | 0.691 | 0.658 | 1.17 | 8.7.E-05 | 0.689 | 0.663 | 1.13 | 1.2.E-06 |
| rs78885399 | 12 | 121178898 | ACADS | T/C | 0.87 | 0.82–0.92 | 9.80.E-09 | 0.172 | 0.202 | 0.797 | 6.5.E-06 | 0.194 | 0.212 | 0.890 | 5.9.E-05 |
Note: JPN, Japanese population; SCZ, schizophrenia; GWAS, Genome-Wide Association Study; REP, replication; SNP, single nucleotide polymorphism; CHR, chromosome; BP, base position (hg19); A1, effect allele; A2, non-effect allele; Freq, frequency. Bold values indicate 5 × 10−8.
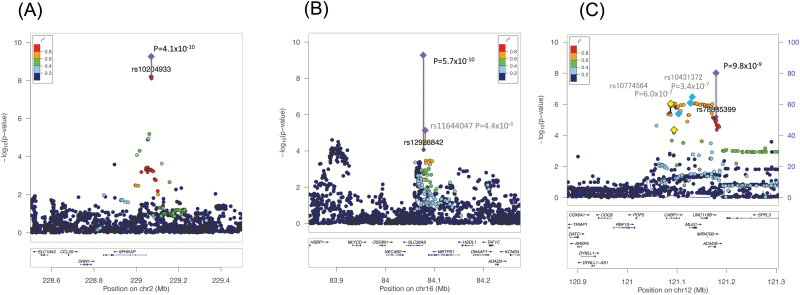
To validate the top signals of the JPN GWAS, we conducted a replication analysis of the SNPs with a P-value of <1 × 10−4 in the discovery GWAS; 178 of 271 SNPs exhibited the same direction of effect (sign test P = 2.7 × 10−7), and 32 SNPs (~12%) showed P < .05, with the same direction of effect in the replication analysis (supplementary table S4). Furthermore, to combine the results of the discovery JPN-SCZ GWAS with the JPN-SCZ REP, a meta-analysis was carried out for increasing the statistical power. Three regions showed a significant association with genome-wide significance (supplementary table S5); the top hit was seen in SPHKAP, as mentioned above (P = 4.1 × 10−10; table 1 and figure 2), and the second top hit was seen at the SNPs around SLC38A8 (rs12928842, P = 5.7 × 10−10; table 1 and figure 2). The last locus was the region between CABP1 and ACADS (rs78885399, P = 9.8 × 10−9; table 1 and figure 2). All of these were novel loci, and previous large PGC-SCZ data did not show marginal P-values for SLC38A8 and CABP1/ACADS (supplementary figure S3). However, in SPHKAP, where a different association peak was observed around the 5′ region of this gene, neighboring loci with LD might contain susceptibility variants for SCZ (supplementary figure S3) because the PGC-SCZ EUR sample showed genome-wide significance (rs7601312, P = 4.7 × 10−8), but the PGC full set (containing extra EAS or AA samples: P = 6.4 × 10−8) showed decreased values (NOTE: after including the replication sample in the PGC article,1 this region was not highlighted as a significant locus for SCZ). The detailed PGC-SCZ EUR results for the index and neighboring SNPs are shown in supplementary table S6.
Fig. 2.
Regional plots of the top hit in the association results based on the meta-analysis of JPN-SCZ GWAS and JPN-SCZ replication data. The Y-axis is −log10 (P-values) of case-control association test of the SNPs, and the X-axis is the chromosomal position (hg19). The linkage disequilibrium (R2) between the top and remaining SNPs is indicated by color. (A) SPHKAP, (B) SLC38A8, and (C) CABP1-ACADS. JPN, Japanese population; SCZ, schizophrenia; GWAS, Genome-Wide Association Study; SNPs, single nucleotide polymorphisms.
Meta-analysis of the JPN (JPN-SCZ GWAS) and PGC-SCZ Results
To increase the sample size, a further meta-analysis was conducted by combining JPN-SCZ GWAS (not included JPN-SCZ REP) and PGC-SCZ (EUR only, because the full set of PGC-SCZ population included some SCZ cases [n = 243]13 used in this study, thereby leading to the duplication of examined datasets1). Manhattan and quantile–quantile plots are shown in supplementary figure S4. Most of the significant SNPs detected in the PGC-SCZ dataset (128 SNPs: 3 SNPs were located in the X chromosome and 125 SNPs in the autosomal chromosome) remained significant, and 7 loci were identified as additional susceptibility genes for SCZ after merging our dataset (supplementary table S7). Among them, a previous study, which applied an identical scheme to ours (ie, meta-analysis of the data from their study and those of the PGC), demonstrated that 2 SNPs were linked to SCZ with genome-wide significance (rs28730912 in TOMM22P6 and rs9398171 in FOXO3)26; thus, 5 novel loci were identified (SFXN5, FHIT, OTOL1, LIN28B, and FLJ35282; table 2 and supplementary figure S5).
Table 2.
Meta-analysis Between JPN-SCZ GWAS and PGC-SCZ EUR
| Meta-analysis (JPN-SCZ GWAS+PGC- SCZ EUR) | JPN-SCZ GWAS | PGC-SCZ EUR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | BP | Closest Gene | A1/A2 | OR | P-value | Case Freq. (A1) | Control Freq. (A1) | OR | P-value | OR | P-value |
| rs62148129 | 2 | 73204203 | SFXN5 | G/A | 0.921 | 2.19E-08 | 0.620 | 0.636 | 0.925 | 3.95E-02 | 0.921 | 1.94E-07 |
| rs1353545* | 3 | 60287845 | FHIT | C/G | 1.07 | 5.82E-09 | 0.706 | 0.683 | 1.10 | 2.16E-02 | 1.06 | 6.76E-08 |
| rs308693 | 3 | 161485407 | OTOL1 | G/A | 1.07 | 1.75E-08 | 0.854 | 0.840 | 1.16 | 1.53E-02 | 1.06 | 1.41E-07 |
| rs28730912** | 3 | 161779956 | TOMM22P6 | T/C | 1.07 | 9.20E-09 | 0.550 | 0.533 | 1.08 | 4.50E-02 | 1.07 | 6.99E-08 |
| rs314272* | 6 | 105462004 | LIN28B | A/G | 0.944 | 4.18E-08 | 0.706 | 0.721 | 0.922 | 4.18E-02 | 0.946 | 2.93E-07 |
| rs9398171** | 6 | 108983527 | FOXO3 | T/C | 0.938 | 2.13E-08 | 0.734 | 0.754 | 0.907 | 1.91E-02 | 0.941 | 2.53E-07 |
| rs9695226* | 9 | 22759396 | FLJ35282 | A/G | 0.941 | 4.46E-08 | 0.691 | 0.692 | 0.958 | 2.75E-01 | 0.939 | 7.30E-08 |
Note: JPN, Japanese population; SCZ, schizophrenia; GWAS, Genome-Wide Association Study; PGC, Psychiatric GWAS Consortium; EUR, European population; SNP, single nucleotide polymorphism; CHR, chromosome; BP, base position (hg19); A1, effect allele; A2, non-effect allele; Freq, frequency. Bold values indicate 5 × 10−8.
*listed in Goes et al but P > 5E-8, **listed in Goes et al and P < 5E-8.
Meta-analysis of the JPN (JPN-SCZ GWAS), Chinese-SCZ GWAS (With Replication for the Top Hit) and PGC-SCZ Results
A recent GWAS of the Chinese population reported 30 novel loci associated with SCZ.18 We conducted further meta-analyses to detect additional susceptibility genes by merging (a) our JPN-SCZ GWAS and the results of the Chinese GWAS/REP data (CHN-SCZ GWAS/REP) and (b) JPN-SCZ GWAS, CHN-SCZ GWAS, and PGC-SCZ EUR (Note: The meta-analysis of each dataset was optimal to obtain precise results. However, we can access only results from the meta-analysis [CHN-SCZ GWAS/REP and PGC-SCZ EUR] for the whole-genome SNPs. Therefore, for the significant SNPs detected by “Chinese only” samples, we only listed the results published in the article by Li et al.)
Based on our meta-analysis between Japanese and Chinese GWASs (JPN-SCZ GWAS and CHN-SCZ GWAS), we identified one novel region around Mir4432 (P = 1.0 × 10−8; table 3; supplementary table S8; supplementary figure S5).
Table 3.
Meta-analysis Between JPN-SCZ GWAS, CHN-SCZ GWAS/REP and PGC-SCZ EUR
| SNP | CHR | BP | Closest Gene | Dataset (JPN-SCZ GWAS Plus) | A1/A2 | Meta all (JPN-SCZ GWAS + CHN-SCZ GWAS + PGC-SCZ EUR) | JPN-SCZ GWAS + CHN-SCZ GWAS/REP | PGC-SCZ EUR | CHN-SCZ GWAS/REP | JPN-SCZ GWAS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | P-value | OR | P-value | OR | P-value | OR | P-value | OR | P-value | ||||||
| rs359260 | 2 | 60486936 | MIR4432 | CHN-SCZ GWAS+ CHN-SCZ REP | G/T | 1.12 | 1.57E-03 | 1.13 | 9.96E-9 | 1.03 | 8.14E-03 | 1.12 | 6.60E-07 | 1.16 | 3.79E-03 |
| rs9970077 | 1 | 177,760,918 | SEC16B | CHN-SCZ GWAS +PGC-SCZ GWAS | A/G | 1.07 | 4.86E-08 | - | - | 1.08 | 5.26E-07 | 1.02 | 3.37E-01 | 1.07 | 7.15E-02 |
| rs12139672 | 1 | 190,940,627 | LOC440704 | CHN-SCZ GWAS +PGC-SCZ GWAS | A/G | 0.941 | 3.85E-08 | - | - | 0.940 | 4.21E-07 | 0.968 | 9.73E-02 | 0.963 | 3.19E-01 |
| rs4833558 | 4 | 118,919,497 | NDST3 | CHN-SCZ GWAS +PGC-SCZ GWAS | T/C | 0.946 | 4.32E-08 | - | - | 0.954 | 5.66E-05 | 0.959 | 3.92E-02 | 0.963 | 3.90E-01 |
| rs12541020 | 8 | 4,817,592 | CSMD1 | CHN-SCZ GWAS +PGC-SCZ GWAS | T/C | 1.06 | 5.58E-09 | - | - | 1.05 | 6.96E-05 | 1.05 | 3.87E-03 | 1.09 | 1.42E-02 |
| rs1864774 | 11 | 83,199,276 | DLG2 | CHN-SCZ GWAS +PGC-SCZ GWAS | T/C | 0.948 | 1.64E-08 | - | - | 0.949 | 2.37E-07 | 0.968 | 7.43E-02 | 0.943 | 1.18E-01 |
| rs12911832 | 15 | 58,985,904 | ADAM10 | CHN-SCZ GWAS +PGC-SCZ GWAS | A/T | 1.06 | 3.78E-08 | - | - | 1.06 | 8.38E-06 | 1.07 | 4.46E-03 | 1.17 | 4.16E-03 |
Note: JPN, Japanese population; GWAS, Genome-Wide Association Study; REP, replication; PGC, Psychiatric GWAS Consortium; EUR, European population; SNP, single nucleotide polymorphism; CHR, chromosome; BP, base position (hg19); A1, effect allele; A2, non-effect allele; SCZ, schizophrenia.
The subsequent meta-analysis between the JPN-SCZ GWAS and the CHN-SCZ GWAS plus PGC-SCZ EUR revealed that 6 additional SNPs associated with genome-wide significance (closest genes: SEC16B, LOC440704, NDST3, CSMD1, DLG2, and ADAM10; table 3; supplementary table S9; supplementary figure S5). Although the results of the JPN-SCZ GWAS exhibited a nominally significant association in some SNPs, the contribution of this significance is primarily derived from the results based on the PGC-SCZ EUR.
PRS Analysis
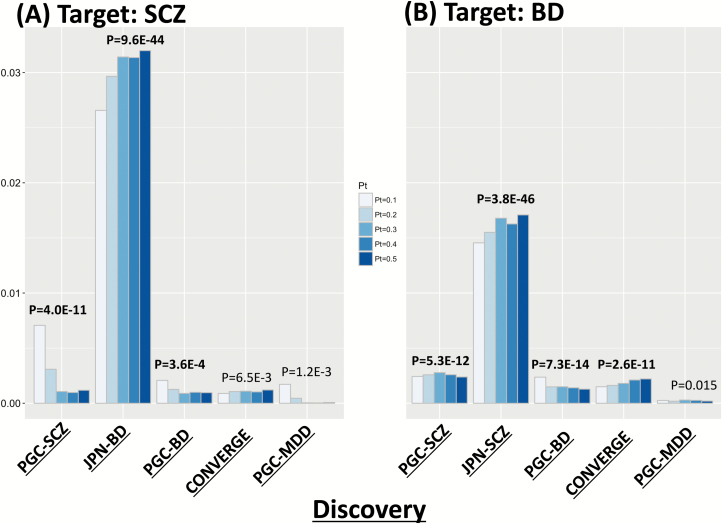
To examine the polygenic architecture of SCZ that is represented by numerous “risk” SNPs, we conducted an PRS analysis using several publicly available datasets. First, as a “trans-population” analysis, we set PGC-SCZ EUR as a discovery dataset, which defined the “risk” SNPs, and JPN-SCZ GWAS as a target dataset (discovery/target: PGC-SCZ EUR/JPN-SCZ GWAS pairs). In this analysis, higher PRS was observed in cases than in controls, with explained variance of approximately 0.7% (P-value of ~10−11), suggesting that shared genetic components of SCZ exist between JPN and EUR populations (figure 3 and supplementary table S10).
Fig. 3.
Polygenic risk score (PRS) analysis. Y-axis indicates the explained variance (Nagelkerke’s R2). (A) Target: JPN-SCZ GWAS (1940 SCZ vs 7408 controls) and (B) Target: JPN-BD GWAS (maximum number: 2964 BD vs 61887 controls, Ikeda et al4). Note that discovery and target samples are independent. BD, bipolar disorder; JPN, Japanese population; SCZ, schizophrenia; GWAS, Genome-Wide Association Study.
Trans-disease PRS analysis of the comparison with JPN-SCZ GWAS revealed that the highest explained variance was observed between the discovery/target JPN-BD GWAS/JPN-SCZ GWAS pair (R2 of ~3%, P-value of ~10−40: figure 3 and supplementary table S10). Another significant PRS was found between the discovery/target PGC-BD/JPN-SCZ GWAS pair (R2 of ~0.2%, P-value of ~3.6 × 10−4; figure 3 and supplementary table S10).
For comparison, PRS was carried out with JPN-BD GWAS as a target dataset. Interestingly, most of the comparisons showed significant PRS, except with PGC-MDD as a discovery dataset. It is reasonable that the lowest P-value was obtained with a comparison between the discovery/target JPN-SCZ GWAS/JPN-BD GWAS pair (R2 of ~1.7%, P-value of ~3.8 × 10−46) because of the larger sample size in JPN-BD GWAS compared with that in JPN-SCZ GWAS. In addition, all the trans-disease/population analyses of JPN-BD GWAS as the target showed larger variance-explained with smaller P-values compared with those of JPN-SCZ GWAS as the target (figure 3 and supplementary table S10).
LD Score Regression Analysis and Genetic Correlation of SCZ with Mood Disorders within/across Population(s)
First, we calculated the SNP heritability of JPN-SCZ GWAS (1025959 SNPs) by the LD score regression method, and the total liability scale (h2) was 0.419 (SE = 0.0571), which was larger than that of JPN-BD reported previously (903223 SNPs, h2 = 0.148, SE = 0.0288) and that of PGC-SCZ EUR (1185779 SNPs, h2 = 0.270, SE = 0.0106).
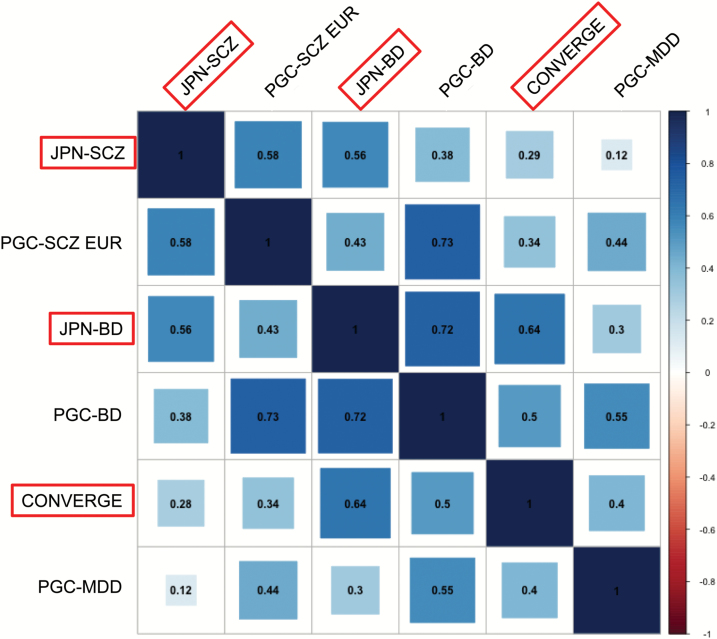
To support the PRS analysis where a shared genetic risk exists across diseases or populations, we calculated genetic correlations among SCZ, BD, and MDD within the same population (using LDSC10) and across populations (using Popcorn12).
First, we examined the genetic correlation of SCZ between the JPN and EUR populations and found a substantial genetic correlation at ~0.58/0.53 for ρge/ρgi, respectively (figure 4, supplementary figure S6 and supplementary table S11). Next, trans-disease correlation was calculated between JPN-SCZ GWAS and BD4 in Japan or MDD (CONVERGE)7 in the Chinese population. The LD score regression estimated the genetic correlations (rg) (a) between SCZ and BD4 in EAS (JPN) to be ~0.56 and (b) between SCZ and MDD7 in EAS (JPN and Chinese) to be ~0.29 (figure 4, supplementary figure S6, and supplementary table S11). Interestingly, we observed that the genetic correlation (rg) between BD4 and MDD7 within EAS was of the same magnitude (~0.64) as that between SCZ and BD. Furthermore, in the EUR samples, the rg among SCZ,1 BD,3 and MDD8 in PGC datasets revealed a similar (but slightly larger) trend to those observed in EAS: SCZ and BD (rg = 0.73), SCZ and MDD (rg = 0.44), BD and MDD (rg = 0.55; figure 4, supplementary figure S6, and supplementary table S11).
Fig. 4.
Genetic correlation of the trans-population and/or trans-disease analysis. The red diagonal line indicates East Asian (EAS) samples. Numerical values indicate estimates of the genetic correlations (rg and σ for “genetic effect”).
Discussion
In this study, we identified 3 novel SCZ susceptibility loci based on the results from JPN samples only (SLC38A8, SPHKAP, and CABP1-ACADS), and 12 novel loci after merging with the PGC-SCZ dataset and/or CHN-SCZ GWAS (SFXN5, FHIT, OTOL1, LIN28B, FLJ35282, MIR4332, SEC16B, LOC440704, NDST3, CSMD1, DLG2, and ADAM10).
A novel gene with the highest association with SCZ based on the Japanese samples was SPHKAP on chromosome 2. This gene encodes a family of A-kinase anchor proteins, which interact with a protein kinase,27 and the function may be related to an abnormality of the signal transduction which is involved in SCZ development, although no functional evidence verifies the correlation between SPHKAP and SCZ. Thus, further studies are warranted to elucidate the functional relevance of this gene and their association with SCZ.
The second gene was SLC38A8, which encodes a sodium-coupled neutral amino acid transporter family; this gene is widely expressed in the brain and plays a role in transporting a broad range of amino acids, including glutamine.28 With the evidence revealing that glutamate abnormality may be related to SCZ,29,30 this gene is also considered to be a promising susceptibility gene for SCZ.
We found the last locus on chromosome 12 (121.1–121.2 Mb); this region contains several genes such as CABP1, MLEC, LINC119B, MIR4700, and ACADS. Hence, highlighting the susceptibility genes among these genes is challenging. However, CABP1 plays a vital role in regulating the gating of voltage-gated calcium ion channels and associates with CAMK2 (calmodulin binding to Ca2+/calmodulin-dependent kinase II)31 and CACNA1C (Ca[V]1.2 channel α[1] subunit).32 In particular, given that CACNA1C is a definitive BD (or possibly SCZ) susceptibility gene, if CABP1 is a genuine susceptibility gene, then abnormal calcium signaling contributes to the development of SCZ, similar to BD.3
In addition, 12 novel SNPs were suggested as possible “common” susceptibility SNPs between JPN, EUR, and/or CHN samples; 5 genes (SFXN5, FHIT, OTOL1, LIN28B, and FLJ35282) after merging our data with PGC-SCZ datasets and additional 7 genes (MIR4432, SEC16B, LOC440704, NDST3, CSMD1, DLG2, and ADAM10) by adding Chinese GWAS results exhibited genome-wide significance. Further meta-analysis is essential to conclude the susceptibility SNPs, as we could not determine functional annotations for these SNPs in this study.
The PRS analysis revealed shared trans-populations/diseases genetic components; it showed that shared genetic risk of SCZ exists between JPN and EUR populations (trans-population PRS analysis for SCZ), but with modest contribution (best R2 of ~0.7%). In contrast, the trans-disease PRS analysis in the JPN population (a comparison between JPN-SCZ and JPN-BD GWAS findings) showed a higher contribution (2%–3% in the discovery/target JPN-SCZ GWAS/JPN-BD GWAS pair or vice versa), corroborating the previous results of the PGC Cross-Disorder Group.9 Also, it is of note that, through the “trans-disease within the same ancestry” analysis of EAS, SCZ/MDD or BD/MDD comparisons did not exhibit significant P-values, which varied from the results of the PGC Cross-Disorder Group;9 this may be attributed to the smaller sample size of MDD among the Chinese population (~5000 cases and ~5000 controls).7
In addition, the genetic correlations analysis supported our PRS results; most of our results for various pairs corresponded to those observed in the PRS analysis. Among these, it is of note that we observed a smaller genetic correlation (rg) between SCZ and BD in the EAS (~0.58) than that in the EUR (~0.73) population. Although our sample size was too small to obtain concrete results, based on the results, we speculate that the higher proportion of the BD type II in our samples (~47%: ie, PGC-BD ~11%3) might influence on this result; a recent study suggested that the genetic component of BD type I was closer to that of SCZ (ie, higher genetic correlation), whereas a lower genetic correlation was observed between BD type II and SCZ, rather higher between BD type II and MDD.33
The current study has several limitations pertinent to the interpretation of the results. First, the control group was not screened for psychiatric conditions, although we believe that this is reasonable given the low prevalence of SCZ (~1%). Therefore, our controls have potential biases about this given other psychiatric conditions are present in the population that may share a genetic component, and this could be an influential design issue. Second, our sample was not large enough to obtain precise results for SNP-based association, PRS, and genetic correlation analyses. Thus, additional samples from the JPN population are required. The power analysis suggested that the discovery samples had only 4 × 10−6~5 × 10−5 power for SCZ to detect a significant association (assuming OR of 1.058, the lowest effect size in the PGC2) of risk with 10%~50% minor allele frequency under an additive model (significance level = 5 × 10−8). Third, different susceptibility genes emerged from EAS or trans-ethnic comparison, and the meta-analysis of the full-set samples was difficult to interpret. Smaller sample size in EAS is a possible confounding factor, however, at least, the precise meta/mega-analysis for EAS sample and the trans-ethnic meta-analysis between EAS and EUR should be re-analyzed, because so far, the whole-SNP results for Chinese only samples were not publicly available. Finally, the results for the PRS and the genetic correlation analyses might be affected by several variables, such as power of the discovery sample, power as a reflection of the sample size of the target sample and LD differences in the populations.
In conclusion, this study indicates that (a) 3 novel loci are associated with SCZ in the JPN population, (b) 12 additional loci are susceptibility genes for SCZ by merging our data with the PGC-SCZ and/or CHN-SCZ GWAS datasets, and (c) SCZ “risk” effect is shared with other psychiatric disorders even across populations.
Funding
This work was supported by the Strategic Research Program for Brain Sciences (SRPBS) from the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP18dm0107097, JP18dm0107087, JP18dm0107100, JP18dm0207005, and JP18dm0107083; part of the BioBank Japan Project from the Ministry of Education, Culture, Sports, and Technology (MEXT) of Japan; GRIFIN of P3GM from AMED under Grant Numbers JP18km0405201 and JP18km0405208; Japan Society for the Promotion of Science (JSPS) Kakenhi Grant Numbers JP25293253, JP16H05378, JP26293266, JP17H04251, and JP16K19785: Grant-in-Aid for Health Labour Sciences Research Grant from the Ministry of Health Labour and Welfare (Comprehensive Research on Disability Health and Welfare); Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from AMED; Grant-in-Aid for Scientific Research on Innovative Areas, “Glial assembly: a new regulatory machinery of brain function and disorders” from MEXT; the Private University Research Branding Project from MEXT.
Supplementary Material
Acknowledgments
We thank all the individuals who participated in the study. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ripke S, O’Dushlaine C, Chambert K, et al. ; Multicenter Genetic Studies of Schizophrenia Consortium; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ikeda M, Takahashi A, Kamatani Y, et al. . A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018;23:639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bigdeli TB, Ripke S, Peterson RE, et al. . Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry. 2017;7:e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyde CL, Nagle MW, Tian C, et al. . Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Converge Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ripke S, Wray NR, Lewis CM, et al. ; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee SH, Ripke S, Neale BM, et al. ; Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown BC, Ye CJ, Price AL, Zaitlen N; Asian Genetic Epidemiology Network Type 2 Diabetes Consortium Transethnic genetic-correlation estimates from summary statistics. Am J Hum Genet. 2016;99:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikeda M, Aleksic B, Kinoshita Y, et al. . Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478. [DOI] [PubMed] [Google Scholar]
- 14. Abecasis GR, Altshuler D, Auton A, et al. ; 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. . Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Chen J, Yu H, et al. . Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49:1576–1583. [DOI] [PubMed] [Google Scholar]
- 19. Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pruim RJ, Welch RP, Sanna S, et al. . LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purcell SM, Wray NR, Stone JL, et al. ; International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goes FS, McGrath J, Avramopoulos D, et al. . Genome-wide association study of schizophrenia in Ashkenazi Jews. Am J Med Genet B Neuropsychiatr Genet. 2015;168:649–659. [DOI] [PubMed] [Google Scholar]
- 27. Kovanich D, van der Heyden MA, Aye TT, van Veen TA, Heck AJ, Scholten A. Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. ChemBioChem. 2010;11:963–971. [DOI] [PubMed] [Google Scholar]
- 28. Hägglund MGA, Hellsten SV, Bagchi S, et al. . Transport of L-glutamine, L-alanine, L-arginine and L-histidine by the neuron-specific Slc38a8 (SNAT8) in CNS. J Mol Biol. 2015;427:1495–1512. [DOI] [PubMed] [Google Scholar]
- 29. Hu W, MacDonald ML, Elswick DE, Sweet RA. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann N Y Acad Sci. 2015;1338:38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howes O, McCutcheon R, Stone J. Glutamate and dopamine in schizophrenia: an update for the 21st century. J Psychopharmacol. 2015;29:97–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamaguchi K, Yamaguchi F, Miyamoto O, et al. . Calbrain, a novel two EF-hand calcium-binding protein that suppresses Ca2+/calmodulin-dependent protein kinase II activity in the brain. J Biol Chem. 1999;274:3610–3616. [DOI] [PubMed] [Google Scholar]
- 32. Oz S, Tsemakhovich V, Christel CJ, Lee A, Dascal N. CaBP1 regulates voltage-dependent inactivation and activation of Ca(V)1.2 (L-type) calcium channels. J Biol Chem. 2011;286:13945–13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stahl EA, Forstner A, McQuillin A, et al. . Genomewide association study identifies 30 loci associated with bipolar disorder. bioRxiv. 2017. doi:https://doi.org/10.1101/173062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.