FIG 2.
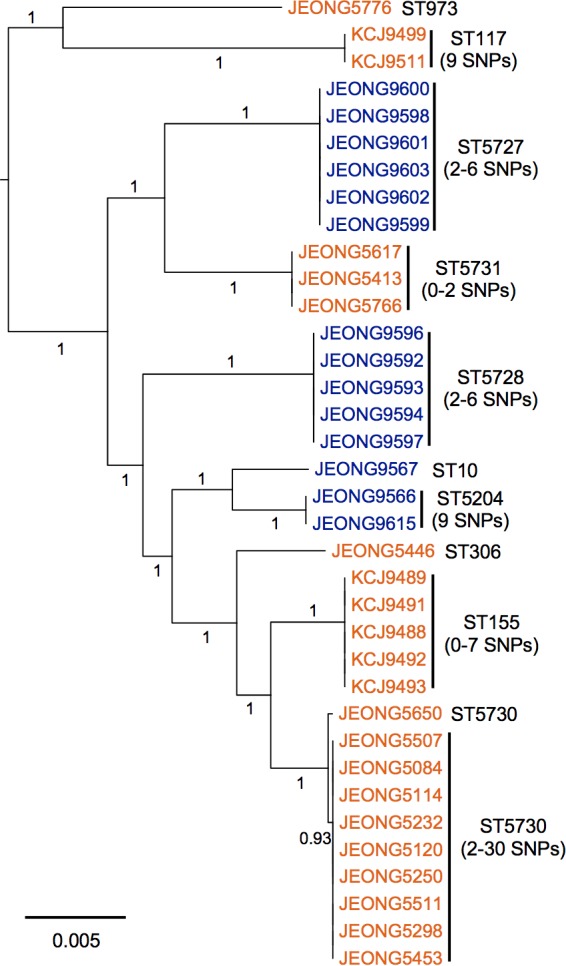
Multidrug-resistant E. coli isolates clustered based on farm location. The maximum-likelihood phylogenetic tree was constructed based on the single-nucleotide polymorphisms (SNPs) identified in the core genomes of the 36 multidrug-resistant E. coli isolates by Parsnp. Of the 36 isolates, 14 were from the NFREC farm (blue) and 22 were from the BRU farm (orange). Sequencing types (ST) of isolates were identified by using the MLST 1.8 software of the Center for Genomic Epidemiology (CGE). The numbers of SNPs were analyzed by using NCBI Pathogen Detection. The numbers listed on the horizontal branches indicate bootstrap values. The scale bar indicates the mean number of nucleotide substitutions per site.
