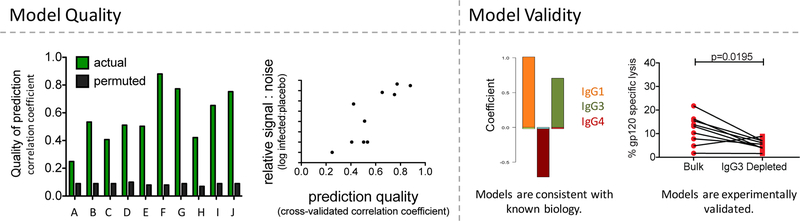
Figure 4. Evaluating model quality and validity.
Left. Prediction quality, as defined by correlation coefficient between cross-validated models learned from actual (green) and permuted (black) data for a set of ten antibody effector functions (A-J). While performance varies considerably (0.2 < r < 0.9) for models learned from real data, permuted data consistently yields essentially random performance (r ~0.1). Relationship between prediction quality and assay signal-to-noise, which suggests that models may be limited by performance characteristics of cell-based assays such as signal to noise (here) and assay reproducibility (50). Right. The biological validity of the models can be supported by prior knowledge, such as the reliance of NK activation predictions antibody types known to have these activities (IgG1 and IgG3) (49), and experimentally confirmed, such as by observing decreased activity when IgG3s are depleted (24).