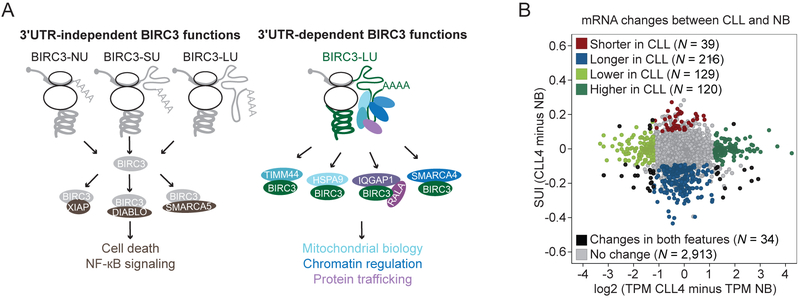
Figure 7. 3′UTR-dependent protein interactors have the potential to be widespread.
(A) 3′UTR-independent BIRC3 functions are mediated by 3′UTR-independent protein interactors (brown), including XIAP, DIABLO, and SMARCA5. They can be accomplished when BIRC3 protein is expressed from transcripts containing no regulatory 3′UTR elements (NU), the short 3′UTR (SU) or the long 3′UTR (LU). 3′UTR-dependent BIRC3 roles include the regulation of mitochondrial biology (light blue), chromatin (dark blue), and protein trafficking (purple). These functions are mediated by BIRC3-LU and the 3′UTR-dependent protein interactors including HSPA9, TIMM44, SMARCA4, IQGAP1, and RALA.
(B) Change in mRNA pattern between CLL4 and NB. 3′UTR ratio change is shown on the y-axis and expression change is shown on the x-axis for genes with more than one 3′UTR. Each dot represents an mRNA. Color-coded upon significant difference between the two samples, calculated as in Figure S1A. Red/blue, increased/decreased expression of the short 3′UTR isoform in CLL; dark/light green, higher/lower expression of mRNA in CLL; black, significant change in mRNA levels and change in 3′UTR isoform ratio, and grey indicates no significant change.