Fig 1.
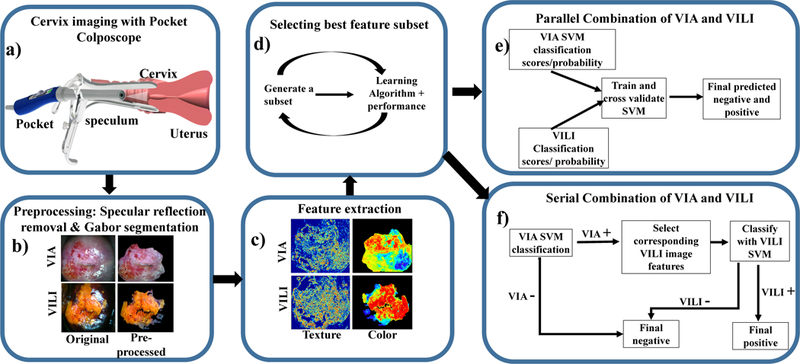
Flow chart of the individual feature extraction and classification processes. a) Image collection: A low-cost, Pocket Colposcope for cervix imaging was used to obtain 134 image pairs of acetic acid (VIA) and Lugol’s iodine contrasts (VILI), b) Pre-processing: These images were preprocessed by applying a specular reflection attenuation algorithm to remove bright white speckles. Custom Gabor filters were applied to the image to select a region of interest for further processing. The pre-processing and segmentation stages were the same for both VIA and VILI, c) Feature extraction: For VIA preprocessed images, Haralick’s texture features, including contrast, correlation, energy and homogeneity, were obtained. Images were also transformed into individual channels of different colorspaces, overall generating 69 features for each VIA image. For VILI images, the same Haralick’s texture and color features were extracted, however approximate lesion stain size was also determined, yielding 70 features, d) A subset of these features were selected with a wrapper feature selection method and selected features were used to train and cross-validate a support vector machine, (e-f) Finally, the features from VIA and VILI were combined using 2 different methods: e) Parallel method and f) Serial method. These were validated to determine improvement of combined classification over using one source of contrast alone.
