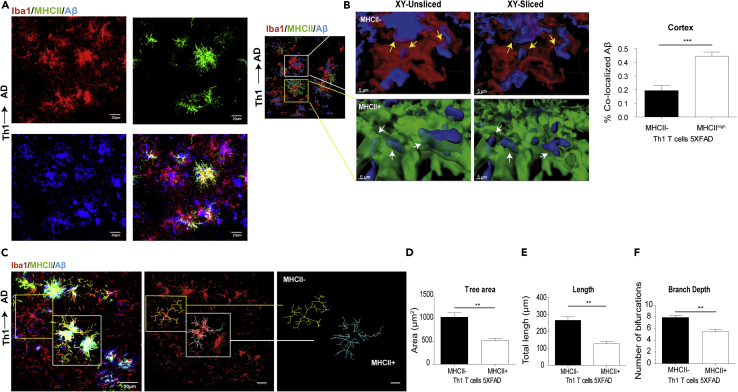
Figure 2.
MHCII+ Cells Are More Phagocytic and Less Ramified than MHCII− Cells in 5XFAD Mice ICV-Injected with Aβ-Th1 Cells
5XFAD mice were ICV-injected with either Aβ-Th1 T cells or PBS, killed at 21 dpi, and their brains collected and analyzed by IHC.
(A) Representative IHC images showing the accumulation of Iba1+ and MHCII + cells at the vicinity of plaque in brain sections derived from Th1→AD mice at 21 dpi and immunolabeled with anti-Iba1 (red), anti-MHCII (green), and anti-Aβ (blue). The rightmost lower panel shows the merged image.
(B) The left image is a 3D reconstruction of z-sections (17.25 μm overall, 0.75 μm/slice), demonstrating the co-localization of Aβ with MHCII− and MHCII+ cells. The images in the middle are higher magnifications of the framed areas, showing MHCII− (top) and MHCII+ (bottom) cells. The images on the right are xy plane slices of the 3D reconstructions. Scale bars, 30 μm (left image) and 5 μm (middle and right images). The yellow and white arrows indicate the co-localization of Aβ with MHCII− and MHCII+ cells, respectively. A quantitative analysis (right graph) of the percentage of Aβ that was co-localized with MHCII− or MHCII + cells in the cortex was conducted with IMARIS. Bars represent means ± SEM.
(C) Representative IHC images showing the backbone of single cells (yellow and white frames indicate MHCII− and MHCII + cells, respectively), traced manually with the IMARIS Filament Tracer plug-in (middle and right images).
(D–F) Quantitative morphometric analyses of randomly selected MHCII+ and MHCII− cortical microglia (layer 2 or 3), performed with IMARIS and statistically analyzed with GraphPad. Bars represent the means ± SEM of the total tree area (D), total branch length (E), and branch depth (F) in one experiment out of four performed (n = 5 mice per group). **p < 0.01, ***p < 0.001 (Student's t test).