Figure 1.
Identification of Transcriptional Regulators of the ESC to EpiLC Transition by Open Chromatin Profiling
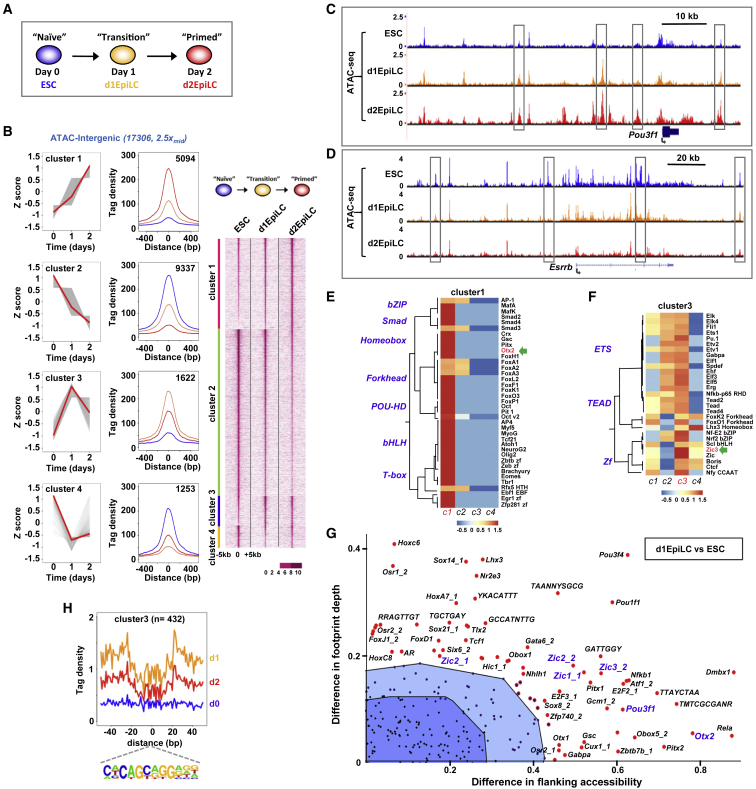
(A) Schematic of the experimental time course of the naive ESC to EpiLC transition.
(B) Heatmap of the ATAC-seq profiles across a 10-kb window of intergenic regions showing > 2.5-fold change in accessibility between any two conditions (right). Average tag densities of each of four identified clusters (middle; blue = ESC, orange = d1EpiLCs, red = d2EpilCs) and average tag density profiles (z scored) are shown across the time course (left). Medians (red) and data for individual peaks (gray) are indicated.
(C and D) University of Santa Cruz (UCSC) genome browser views of the ATAC-seq profiles around the Pou3f1 (C) and Esrrb (D) loci. Dynamically changing peaks are boxed.
(E and F) Heatmap showing the enrichment of transcription factor binding motifs across each of the open chromatin cluster profiles (z-normalized p values) for motifs enriched in cluster 1 (E) or cluster 3 (F).
(G) BaGFoot analysis of the open chromatin regions in ESCs and d1EpiLCs (using all dynamic intergenic peaks from clusters 1 to 4 in B). The top-right quadrant from the whole plot (see Figure S4) is shown. Motifs showing significant increases in either local accessibility and/or footprint depth are labeled. Non-significant regions are shown in the dark (bag) or light (fence) blue-shaded regions.
(H) Average tag densities in a 100-bp window surrounding the ZIC3 binding motif (bottom) in ESCs (blue), d1EpiLCs (orange), or d2EpilCs (red) are shown for ATAC-seq peaks from cluster 3.
See also Figures S1–S5.