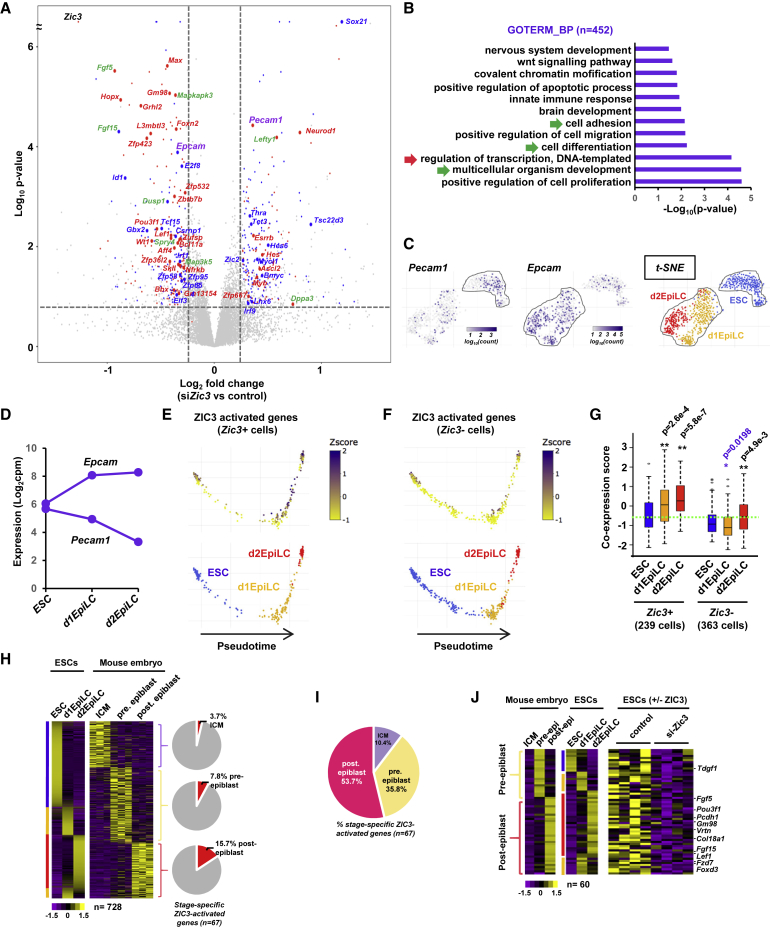
Figure 4.
Identification of Zic3-Regulated Genes
(A) Volcano plot showing changes in gene expression in d1EpiLCs following Zic3 knockdown by small interfering RNA (siRNA). Gene names are color coded for transcription factors (red for direct and blue for indirect targets) and signaling molecules (green). Pecam1 and Epcam are shown in purple.
(B) Gene ontology analysis of all ZIC3-regulated genes for the biological process (BP) category.
(C) Expression of Pecam1 and Epcam at single cell level. Data are plotted as log10 counts per cell, and superimposed on t-SNE analysis of the entire RNA-seq dataset for three time points (right).
(D) Expression of Epcam and Pecam1 from aggregated scRNA-seq analysis in the indicated cell populations and shown as log2 counts per million base pairs (cpm).
(E and F) Pseudotime analysis of ESCs, d1EpiLCs, and d2EpilCs based on the entire scRNA-seq dataset (bottom). The binarized expression (transformed z score) of the ZIC3 activated genes (n = 240) is plotted on top of these profiles (top) in cells that either show Zic3 expression (E) or lack the expression of Zic3 and Otx2 (F). Pseudotime analysis was initially performed with all cells but in each case only cells exhibiting the Zic3 expression characteristics are shown.
(G) Boxplot showing the co-expression scores for the ZIC3-activated genes in ESCs, d1EPiLCs, and d2EpilCs. Cells are split according to whether Zic3 is expressed or not. Horizontal lines represent the median score, and the dotted green line is the median score in ESCs.
(H) Heatmaps showing the expression levels of genes categorized as uniquely expressed in ICM, pre-epiblast, or post-epiblast (right; Boroviak et al., 2015) and the corresponding expression levels in the aggregated scRNA-seq from ESCs, d1EPiLCs, and d2EpilCs (left). The heatmap is sorted based on the scRNA-seq data from the ESC-derived cells at each of the expression clusters from mouse embryos. Data are row z normalized for each dataset. The pie charts show the proportions of each of the stage-specific gene sets that are activated by ZIC3.
(I) Pie chart showing the proportions of ZIC3-activated lineage-specific genes from each stage of embryonic development.
(J) Heatmap showing the effect of Zic3 depletion on the pre- and post-epiblast stage-specific genes in d1EpiLCs (right) and the heatmaps for the corresponding gene expression levels in early embryonic developmental stages (left) or ESCs, d1EpiLCs, and d2EpilCs (center).
Genes shown are from the red quadrants of the bottom two pie charts in (H). All heatmaps are individually z normalized.