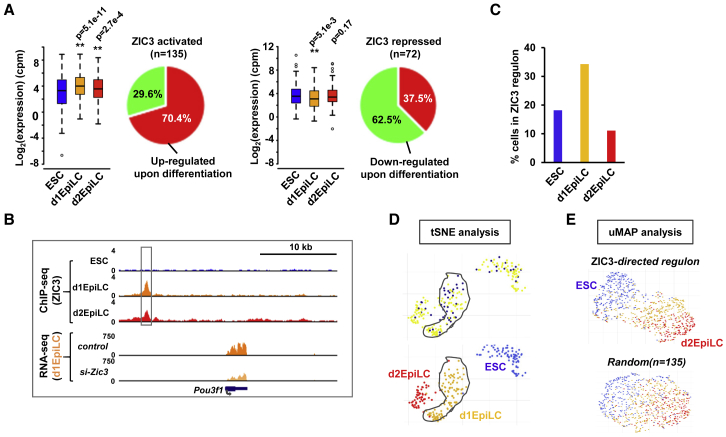
Figure 5.
The Direct ZIC3 Target Gene Network
(A) Boxplots of the expression of directly regulated ZIC3 target genes (i.e., bound by ZIC3) in ESCs, d1EpiLCs, and d2EpilCs for activated (top) or repressed (bottom) genes. The proportions of direct ZIC3 target genes increasing and decreasing expression in d1EpiLCs upon differentiation from ESCs are shown in the pie charts on the right.
(B) UCSC genome browser views of the ZIC3 ChIP-seq (top) and RNA-seq (bottom) profiles around the Pou3f1 locus. The major ZIC3 binding peak is boxed.
(C) AUCell analysis of the expression of the directly activated ZIC3 target gene regulon in ESCs, d1EPiLCs, and d2EpilCs. The percentage of cells is shown from each stage of differentiation that exhibits co-expression of the ZIC3 regulon.
(D) An scRNA-seq analysis of the ZIC3 regulon expression. Data are mapped (blue marked cells; top) on top of tSNE analysis of the entire RNA-seq dataset (bottom), with the originating cell types color coded. The d1EpiLCs are circled. Only Zic3 positive cells are shown.
(E) Uniform manifold approximation and projection (uMAP) analysis of the scRNA-seq ESCs, d1EPiLCs, and d2EpilCs using either the 135 ZIC3-activated direct target genes in the ZIC3 regulon (top) or 135 randomly selected genes (bottom) to drive the clustering. Cells are color coded according to their known origins (blue = ESCs, orange = d1EpiLCs, and red = d2EpiLCs).