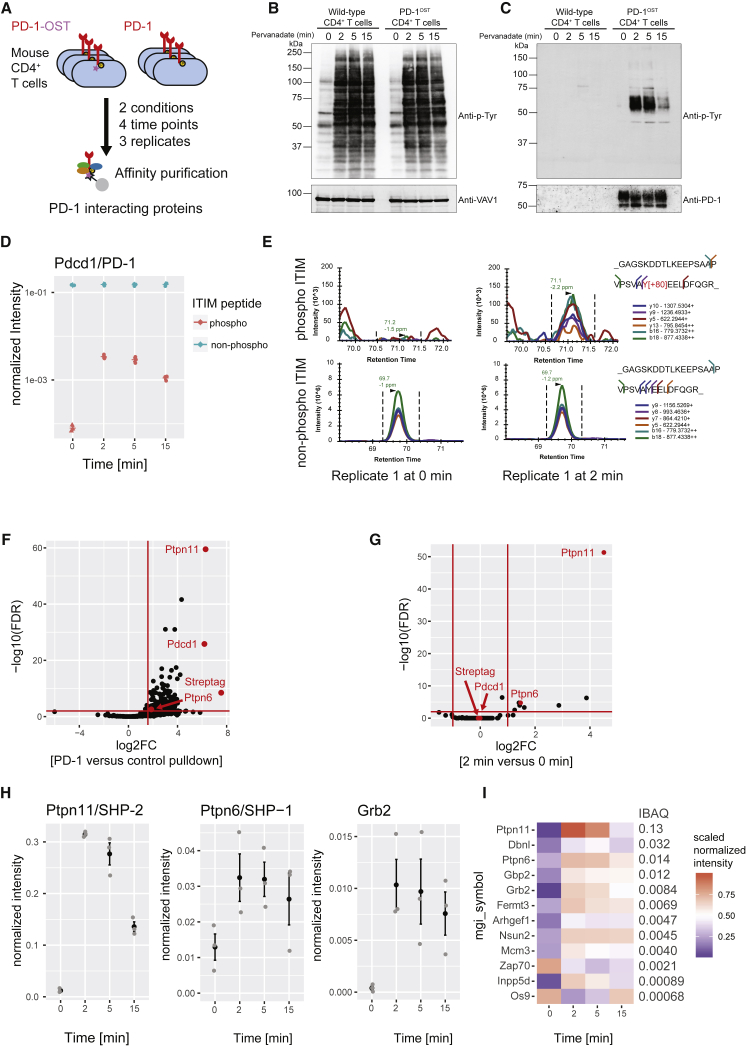
Figure 1.
Composition and Dynamics of the PD-1 Signalosome in Primary CD4+ T Cells
(A) Overview of AP-MS analysis of PD-1+ CD4+ effector T cells from WT mice (PD-1) and gene-edited mice expressing endogenous PD-1 molecules tagged with a Twin-Strep-tag (PD-1OST). T cells were lysed prior to or after stimulation for 2, 5, and 10 min with pervanadate followed by affinity purification of PD-1-OST protein complexes.
(B) Immunoblot analysis of equal amounts of proteins from total lysates of PD-1+ CD4+ effector T cells from WT and PD-1OST mice left unstimulated (0) or stimulated with pervanadate for 2, 5, and 15 min and probed with antibody to phosphorylated tyrosine (Anti-p-Tyr) or anti-VAV1 (loading control).
(C) Immunoblot analysis of equal amounts of proteins from total lysates of cells as in (B), subjected to affinity purification on Strep-Tactin-Sepharose beads, followed by elution of proteins with D-biotin, and probed with antibody to phosphorylated tyrosine (Anti-p-Tyr) or anti-PD-1 (affinity purification control). Left margins of (B) and (C), molecular size in kilodaltons. In (B) and (C), data are representative of three independent experiments.
(D) Intensity values for the non-phosphorylated and phosphorylated forms of the peptide containing the PD-1 ITIM motif. Values were normalized to that of the PD-1 bait (see STAR Methods).
(E) Elution profile of the six different fragment ions from the phosphorylated and non-phosphorylated forms of the ITIM-containing peptide measured at 0 and 2 min after pervanadate treatment.
(F) Volcano plot showing proteins significantly enriched in CD4+ T cells expressing PD-1-OST molecules compared with control CD4+ T cells expressing similar levels of untagged PD-1 molecules at 2 min after pervanadate treatment.
(G) Volcano plot showing proteins significantly enriched in CD4+ T cells expressing PD-1-OST molecules 2 min after pervanadate treatment compared with untreated CD4+ T cells. In (F) and (G), the PD-1/PDCD1, SHP-2/PTPN11, SHP-1/PTPN6 proteins, and Twin-Strep-tag peptide (Streptag) are shown in red, and the x and y axes show the average fold change (log2FC) in protein abundance and the statistical significance, respectively (see STAR Methods).
(H) Intensity for selected interactors across the conditions and replicates. Mean ± SEM is depicted (black), and individual values are shown as gray dots.
(I) Heatmap depicting the average intensity of PD-1 interactors across time points (row-normalized to the maximal value). The iBAQ column shows the stoichiometry of interaction of each prey with the PD-1-OST bait 2 min after pervanadate treatment.