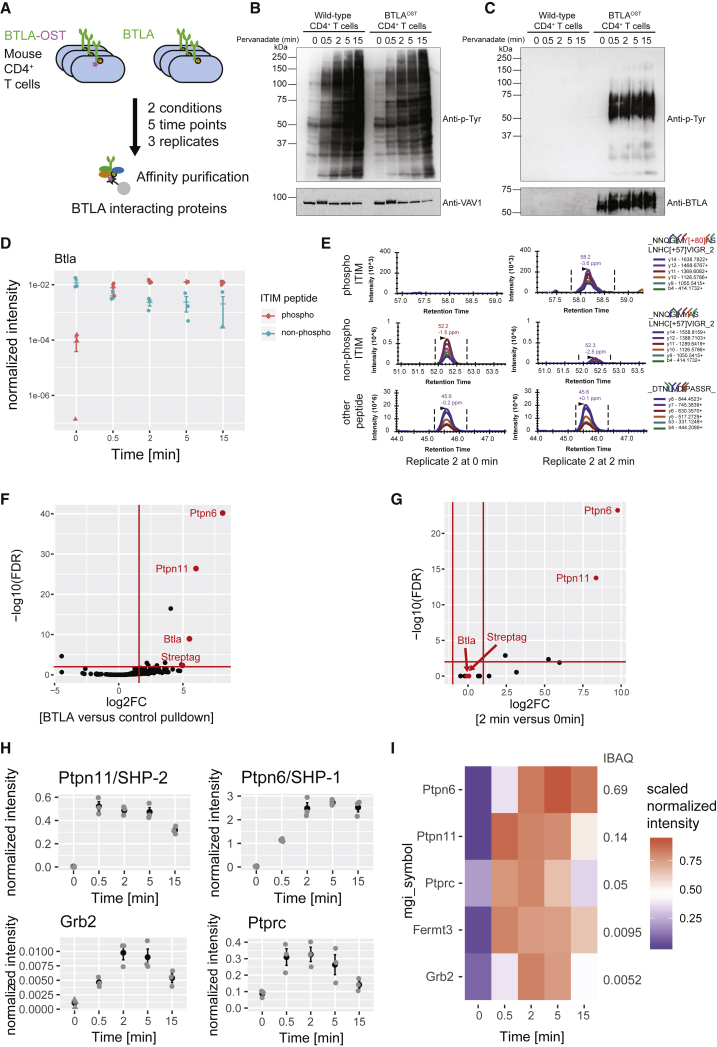
Figure 2.
Composition and Dynamics of the BTLA Signalosome in Primary CD4+ T Cells
(A) Overview of AP-MS analysis of BTLA+ CD4+ effector T cells isolated from WT mice (BTLA) and gene-edited mice expressing endogenous BTLA molecules tagged with a Twin-Strep-tag (BTLAOST). T cells were lysed prior to or after stimulation for 0.5, 2, 5, and 15 min with pervanadate followed by affinity purification of BTLA-OST protein complexes.
(B) Immunoblot analysis of equal amounts of proteins from total lysates of WT and BTLAOST CD4+ T cells left unstimulated (0) or stimulated for 0.5, 2, 5, and 15 min with pervanadate and probed with antibody specific for phosphorylated tyrosine (Anti-p-Tyr) or VAV1 (loading control).
(C) Immunoblot analysis of equal amounts of proteins from total lysates of cells as in (B), subjected to affinity purification on Strep-Tactin-Sepharose beads, followed by elution of proteins with D-biotin, and probed with antibody specific for phosphorylated tyrosine (Anti-p-Tyr) or BTLA (affinity purification control). Left margins of (B) and (C), molecular size in kilodaltons. Data in (B) and (C) are representative of three independent experiments.
(D) Intensity values for the non-phosphorylated and phosphorylated forms of the peptide containing the BTLA ITIM motif. Values were normalized to that of the BTLA bait, and one value has been imputed (triangle).
(E) Elution profile of the six different fragment ions from the phosphorylated and non-phosphorylated form of the ITIM-containing peptide measured at 0 and 2 min after pervanadate treatment.
(F) Volcano plot showing proteins significantly enriched in CD4+ T cells expressing BTLA-OST molecules compared with control CD4+ T cells expressing similar levels of untagged BTLA proteins at 2 min after pervanadate treatment.
(G) Volcano plot showing proteins significantly enriched in CD4+ T cells expressing BTLA-OST molecules 2 min after pervanadate treatment compared with untreated cells. In (F) and (G), the BTLA, SHP-2/PTPN11, SHP-1/PTPN6 proteins, and Twin-Strep-tag peptide (Streptag) are shown in red, and the x and y axes show the average fold change (log2FC) in protein abundance and the statistical significance, respectively.
(H) Intensity for selected interactors across the different replicates. Mean ± SEM is depicted (black), and individual values are shown as gray dots.
(I) Heatmap depicting BTLA interactor intensity across time points (row-normalized to the maximal value). The iBAQ column shows the stoichiometry of interaction of each prey with the BTLA bait 2 min after pervanadate treatment.