Figure 5.
Comparison of the Two Cisplatin-Treated RPF Populations Highlights a Unique Alveolar Lesion Identity
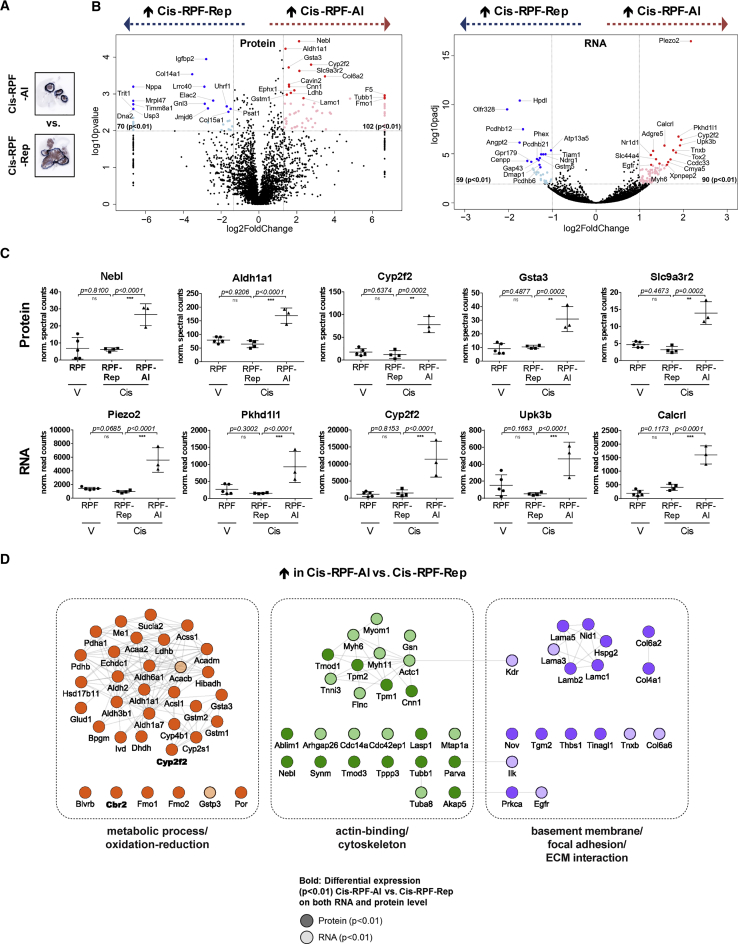
(A) Representative images (CDH1 staining) of samples used for RPF-Al versus RPF-Rep comparison (for images of all three RPF-Al and four RPF-Rep mice analyzed, see Figure S3A).
(B) Most discriminatory genes in RPF-Al versus RPF-Rep comparison (determined using beta-binomial test for protein and DESeq2 for RNA-seq data). All genes with p values < 0.01 and −2.5 ≥ fold change (FC) ≥ 2.5 (protein, left plot) or −2 ≥ FC ≥ 2 (RNA, right plot) are colored in light blue (UP in RPF-Rep) or light red (UP in RPF-Al), respectively. Highlighted with proteins names (and marked with dark blue and red dots) are the 15 most highly differentially expressed genes in each direction (by p value after FC filtering).
(C) Expression plots of the 5 most differential genes on protein (top row) and RNA level (bottom row), respectively (∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001; n.s., not significant).
(D) Highly differentially expressed proteins (p < 0.01 at the protein and/or RNA level) form three highly interactive functional clusters: metabolic process and oxidation-reduction (left), actin-binding and cytoskeleton (middle), and basement membrane, focal adhesion, and ECM interaction (right).