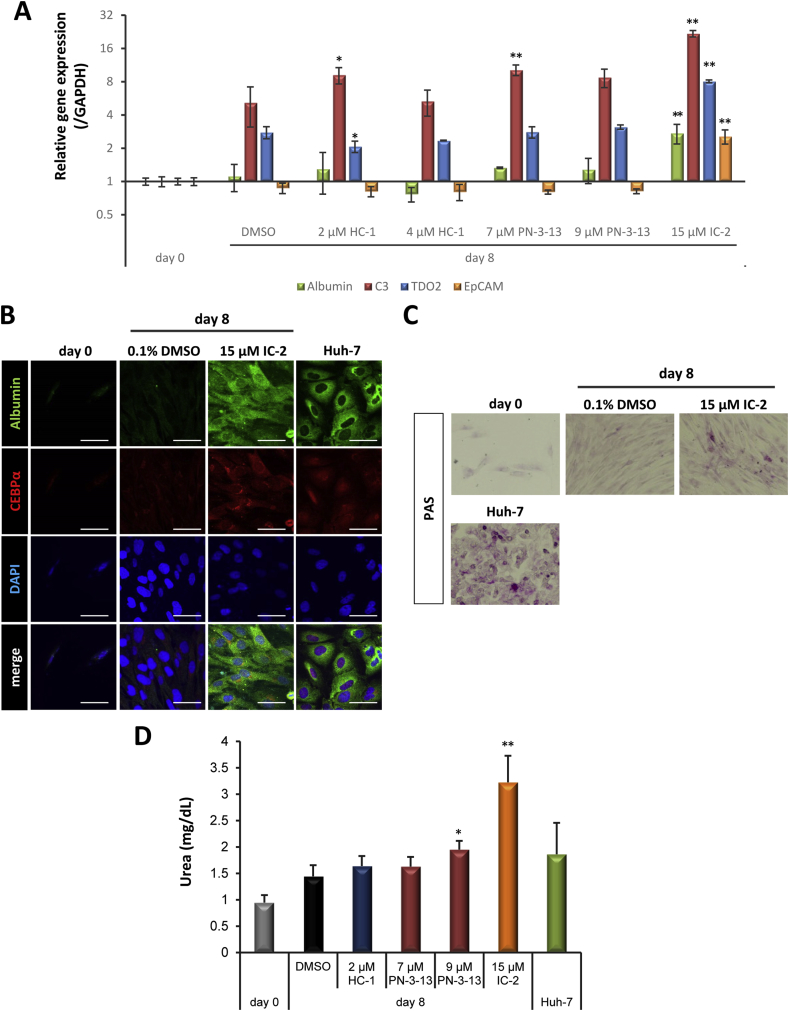
Fig. 5.
IC-2 potently induced hepatic differentiation of UE7T-13 cells. A. Liver-specific gene expression was confirmed by qRT-PCR analysis. Hepatocyte markers (albumin, C3, and TDO2) and a hepatic stem cell marker (EpCAM) were measured. Data are expressed as the means ± SD of three experiments. Analysis by one-way ANOVA, followed by Dunnett's post-hoc test, was used to compare the relative gene expression of UE7T-13 cells following treatment with different compounds at day 8. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO. B. Immunofluorescence microscope image of hepatocyte-specific protein expression. Expression of albumin and C/EBPα was confirmed on day 0 and on day 8. Four different images including albumin and C/EBPα, DAPI and their merge were shown in four different conditions. Scale bar represents 50 μm. C. Glycogen storage in UE7T-13 cells treated with small molecule compounds. PAS staining was performed to demonstrate glycogen storage on day 0 and 8. Huh-7 cells were used as a positive control. D. Urea production in cells treated with small molecule compounds. After incubation of UE7T-13 cells with small molecule compounds for eight days, culture media were fleshly replaced by media including 5 mM ammonium chloride and cells were further incubated for 96 h. Huh-7 cells were used as a positive control. Data are expressed as the means ± SD of six experiments. One-way ANOVA, followed by Dunnett's post-hoc test, was used to compare urea production in UE7T-13 cells following treatment with different compounds at day 8. *P < 0.05 and **P < 0.01 compared with 0.1% DMSO.