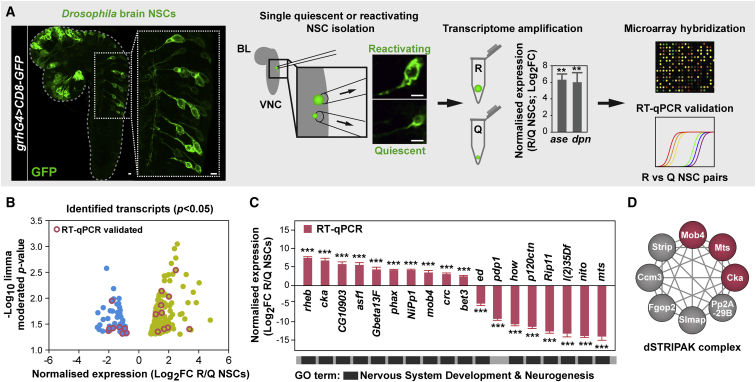
Figure 1.
Single-Cell Transcriptome Analysis of Reactivating NSCs
(A) Workflow: individual quiescent (Q) and reactivating (R) NSCs expressing CD8-GFP driven by grh-Gal4 were harvested from 17 h ALH CNSs, their mRNA reverse transcribed, and resulting cDNA amplified. Quantitative real-time PCRs confirmed higher ase and dpn expression in reactivating versus quiescent NSCs (normalized fold change [log2FC]; n = 3 NSC reactivating/quiescent pairs; error bars: SEMs; Student’s t test, ∗∗p < 0.01). NSC transcriptomes were compared on whole-genome microarrays (reactivating versus quiescent; three pairs) and a subset of identified targets validated by quantitative real-time PCRs. ALH, after larval hatching; BL, brain lobe; VNC, ventral nerve cord. Scale bars: 10 μm.
(B) Distribution of identified transcripts according to average fold change expression (x axis; log2FC) and p value (y axis; limma moderated t test; −log10 p value; p < 0.05). See also Table S1 and Figure S1.
(C) Normalized expression levels in reactivating versus quiescent NSCs obtained by quantitative real-time PCR for a subset of targets (log2FC; n = 3 NSC reactivating/quiescent pairs; error bars: SEMs; Student’s t test; ∗∗∗p < 0.001). The results validate the data from the microarray analysis. Most of the targets selected are classified under “nervous system development” and “neurogenesis” GO terms.
(D) STRING-based interaction network of a Drosophila PP2A-STRIPAK complex reported to inhibit Hippo signaling (Zheng et al., 2017, Liu et al., 2016, Ribeiro et al., 2010), highlighting (pink) the components identified in our transcriptome analysis and functionally characterized in this study.