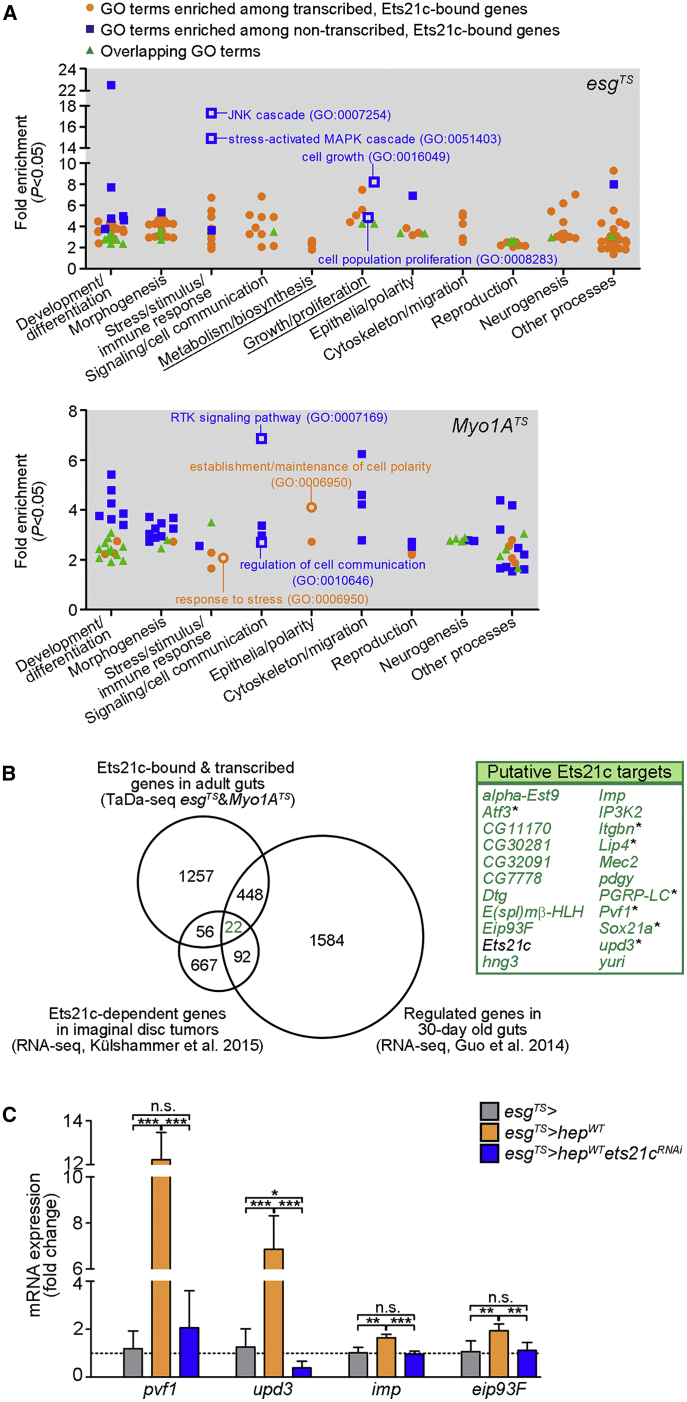
Figure 4.
Ets21c Binds to Actively Transcribed Genes and Those Devoid of PolII Binding
(A) GO analysis of gene loci occupied by Ets21c and PolII in esgTS- and Myo1ATS-expressing adult midgut cells identified by TaDa (see also Tables S2 and S3). Plots depict functional GO terms enriched among the top 250 candidate genes bound by Ets21c. Color coding represents the transcriptional activity based on PolII binding. Each dot represents a single GO term with fold enrichment ≥2 and p < 0.05. Underlined GO groups were overrepresented in esgTS but not Myo1ATS samples.
(B) Venn diagram shows 22 putative Ets21c target genes that overlap between the Ets21c-TaDa dataset of adult midguts and the transcriptomes of Drosophila tumors and aging guts (see also Table S4). Stars indicate genes with reported function in the intestine.
(C) qRT-PCR shows the expression of putative Ets21c target genes pvf1, upd3, imp, and eip93F in 6-day-old midguts overexpressing hepWT and ets21cRNAi in ISCs/EBs (esgTS). Data represent means (SDs); n = 4–5; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. = non-significant.