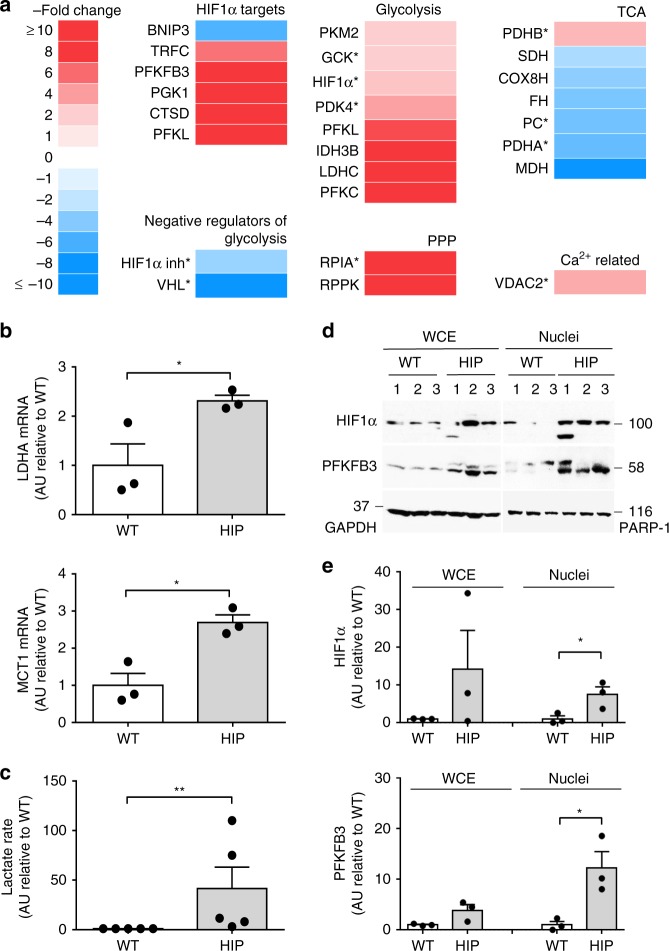
Fig. 1.
hIAPP upregulates the HIF1α/PFKFB3 stress pathway. a Summary of the differentially expressed genes of interest after microarray analysis25 performed on RNA isolated from wild type (WT) and hIAPP overexpressing (HIP) rat islets (4.5 months) presented as a fold change over WT. Asterisk represents genes that were differentially expressed in one experiment. b LDHA and MCT1 mRNA levels in HIP versus WT as measured by qRT-PCR. c Lactate production rate (fold change) measured in isolated islets from HIP relative to WT. d Representative Western blot of PFKFB3 and HIF1α protein levels in whole cell extracts and nuclear-enriched fractions of islets from 6 months old WT (3) and HIP (3) rats. GAPDH and PARP were used as loading controls for whole cell extract (WCE) and nuclear extracts, respectively. e Quantification of HIF1α (upper panel) and PFKFB3 (lower panel) in cytoplasmic and nuclear fractions. Data are presented as mean ± SEM, n = 3 independent experiments for (b) and (d), and n = 4 independent experiments for (c). Statistical significance was analyzed by Student t-test (*p < 0.05, **p < 0.01)