Figure 1.
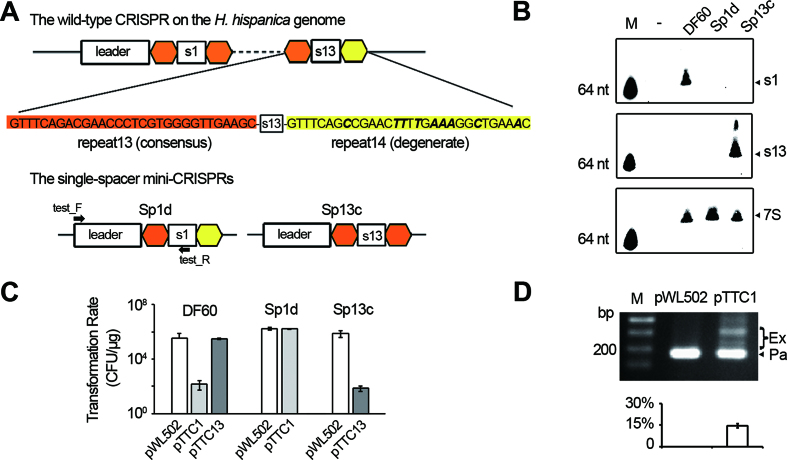
The crRNA production and interference effects were not detected for the terminal spacer. (A) Schematic representation of the WT CRISPR on the H. hispanica genome and two single-spacer mutants. In the WT CRISPR, the most leader-distal spacer (spacer13) is followed by a degenerate repeat (yellow), which differs from the consensus repeat sequence (orange) by 9 nucleotides (in bold and italic). In the mutants Sp1d and Sp13c, spacer1 and spacer13 are followed by the degenerate and the consensus repeat, respectively. Primers employed in panel D are indicated. (B) Northern analysis of the crRNA molecules of spacer1 (s1) and spacer13 (s13) in different strains. 7S RNA was probed as the internal control. M, a biotin-labeled 64-nt ssDNA. -, an empty control without RNA loading. (C) A plot showing the transformation rate of different target plasmids in DF60, Sp1d or Sp13c cells. The target plasmids pTTC1 and pTTC13 are pWL502 derivatives carrying the protospacer of spacer1 and that of spacer13, respectively. The protospacer sequence was designed immediately downstream of a canonical PAM, i.e. 5′-TTC-3′. CFU, colony-forming units. (D) Sp1d cells transformed by pTTC1 were inoculated into the liquid medium, and after 5-day culturing, cells were lysed and subjected to PCR analysis. M, dsDNA size marker. The ∼180 bp PCR products correspond to the parental Sp1d structure (Pa), while the larger-sized products represent those expanded by one or more new spacers (Ex). The plot below the gel shows the percentage of the intensity of ‘expanded’ bands (Ex/(Ex+Pa)). Error bars indicate the standard deviation (SD) calculated from three independent replicates.