Figure 4.
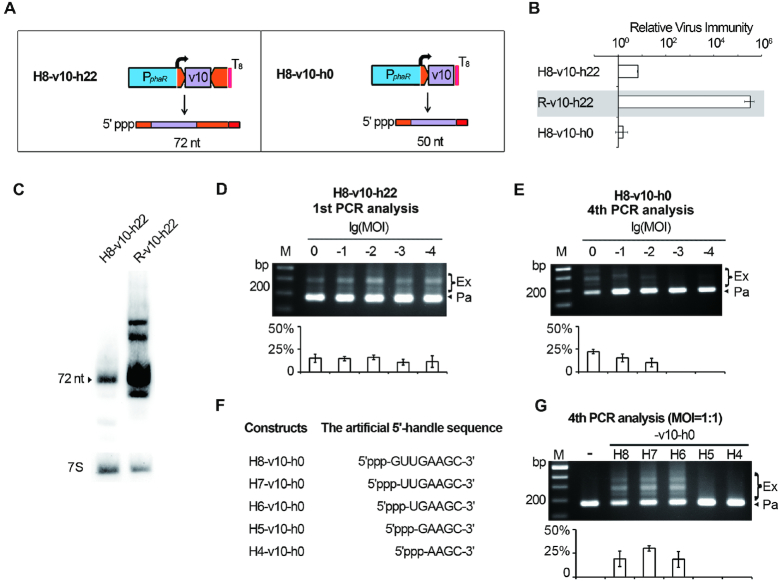
The last 6 nt of 5′ handle is required for priming adaptation to HHPV-2. (A) The H8-v10-h22 and H8-v10-h0 constructs producing Cas6-independent crRNAs (predicted sizes are given). As indicated by the curved arrows, transcription was designed to initiate exactly from the beginning nucleotide of the DNA sequence for 5′ handle. (B) The relative virus immunity (RVI) provided by H8-v10-h22 and H8-v10-h0 in Sp1d cells. Data of R-v10-h22 (against a grey background) is shown again to facilitate comparison. (C) The v10-crRNA from H8-v10-h22 showed a largely decreased stability compared to that from R-v10-h22. 7S RNA was probed as the internal control. Adaptation to HHPV-2 was primed by H8-v10-h22 (readily detected in the first PCR analysis) (D) or H8-v10-h0 (hardly detected until the 4th PCR analysis) (E). (F) Constructs producing crRNAs with a truncated 5′ handle (and no 3′ handle). Note that RNA transcripts from these constructs carry a natural 5′ triphosphate, which may need to be removed prior to Cascade assembly. (G) HHPV-2 adaptation primed by constructs listed in panel F. –, the empty pWL502 control. Adaptation was monitored as illustrated in Figure 3A, and infection was performed at the specified MOIs. The plots show the percentage of the intensity of ‘expanded’ bands (Ex/(Ex+Pa)) for each gel lane. Error bars indicate the SD calculated from three independent replicates.